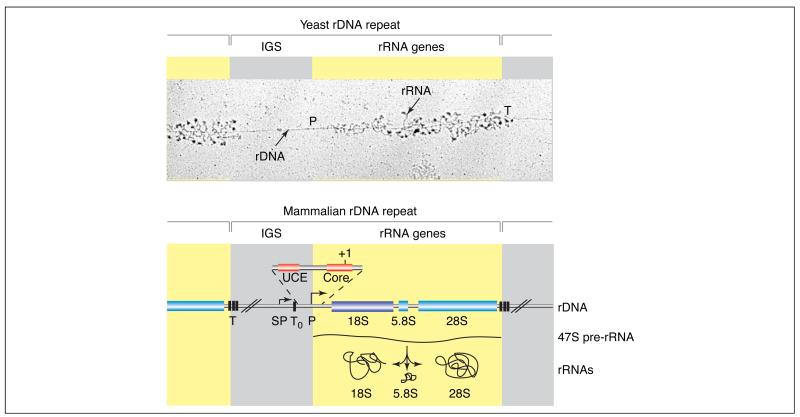
Figure 1. Organization of the rRNA genes.
The repetitive nature of the rDNA is illustrated in an electron microscopic image of a yeast nuclear chromatin spread (‘Miller spread’). Progressively longer rRNAs (stained for associated proteins) emanate from the many pol I complexes as they transcribe the rDNA, beginning at the promoter (P) and finishing at the terminator (T). Beneath, a representative mammalian rDNA repeat is outlined (not drawn to scale). Each human rDNA repeat unit (GenBank accession number: U13369) of ~43 kb contains an intergenic spacer (IGS) of ~30 kb (grey), which contains the transcription regulatory elements, and ~13 kb of sequences encoding the precursor rRNA (yellow). The rRNA genes are present in a single transcription unit, transcribed by pol I to yield a 47S precursor rRNA that is, in part, co-transcriptionally processed and modified by methylation and pseudo-uridinylation to produce the mature 18S, 5.8S and 28S rRNAs. Pol I initiates transcription at the human rDNA promoter (P), which contains an essential core element from −45 to +18 relative to the start site (+1), and an upstream control element (UCE) from −156 to −107 [7]. A spacer-promoter (SP) upstream of the gene promoter directs pol-I-dependent transcription of short-lived transcripts of unknown function. Several transcription-termination elements are located at the 3′ end of the transcribed region of the rRNA genes (T) and immediately upstream of the rRNA gene transcription start site (T0), between the spacer and gene promoters [3].