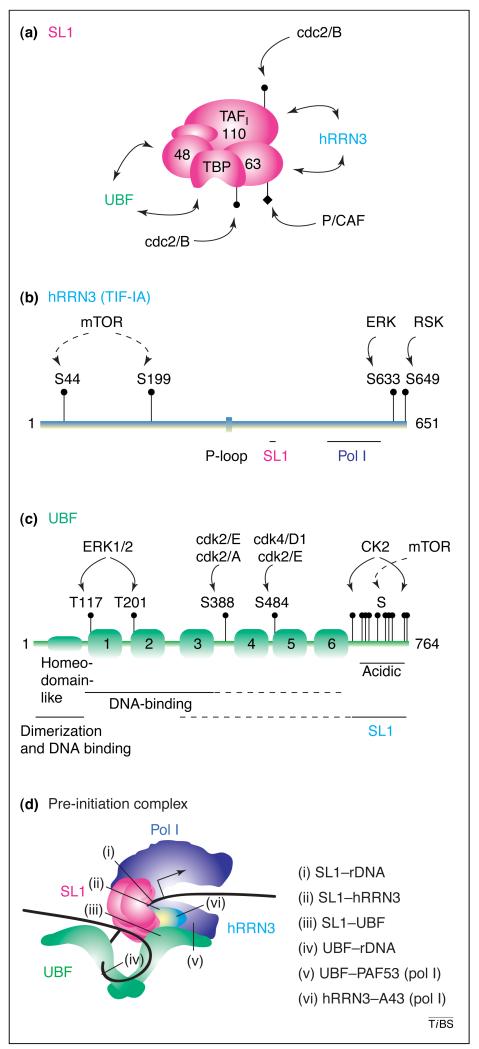
Figure 2. The molecular architecture of the mammalian pol I transcription factors, modification sites and interactions in the transcription pre-initiation complex.
(a) Selectivity factor SL1 is an ~300-kDa complex of the TATA-binding protein (TBP) and at least three pol-I-specific TBP-associated factors (TAFIs) of 110, 63 and 48 kDa (GenBank accession numbers: NM_003194, NM_005679, NM_005680 and NM_005681). The mouse complex is TIF-IB and the homologous TAFIs are 95, 68 and 48 kDa, respectively (GenBank accession numbers: Y09974, Y09973 and Y09972). The SL1 subunits that interact with UBF and hRRN3 and post-translational modifications of TIF-IB [cdc2-cyclin B-mediated phosphorylation and p300/CBP-associated factor (P/CAF) acetylation] are indicated. (b) Human RRN3, a 651-amino acid polypeptide (GenBank accession number: NM_018427), and mouse homologue TIF-IA have sequence and functional homology to Saccharomyces cerevisiae Rrn3p. Conserved regions important for the interactions with SL1 (411–415) and Pol I (512–609) [18] and an ATP/GTP-binding site motif (P-loop; 333–340), of as yet unknown function, are indicated. TIF-IA activity can be regulated by the mTOR kinase and ERK-MAPK signalling pathways (ERK and RSK). (c) Human upstream binding factor UBF (also referred to as UBF1; GenBank accession number: NM_014233) is a 764-amino acid polypeptide of 97 kDa, which is highly conserved in vertebrates and is an activator of pol I transcription. The N-terminal domain is involved in dimerization (essential for the transcription activation function of UBF) and contains a homeo-domain-like motif (possibly involved in DNA binding). In addition, the protein has six high-mobility group (HMG) boxes (labelled 1–6). HMG boxes 1, 2 and 3 are involved in DNA binding through the minor groove. The C terminus, which contains 77% acidic amino acids and is rich in serine residues, can interact with SL1 and, in addition, might influence the binding of HMG box 1 to DNA in a UBF dimer. Human UBF2 is a 94-kDa protein of unknown function encoded by the same gene as UBF1, and is identical to UBF1 except for its lack of 37 amino acids from HMG box 2 due to alternative splicing [80]. Post-translational modifications of UBF include acetylation and phosphorylation via PI3K, cyclin-dependent kinases, the ERK–MAPK pathway and the mTOR pathway [signalling via p70 ribosomal S6 kinase to serine residues in the C terminus, also shown to be consensus phosphorylation sites for Casein Kinase 2 (CK2)]. (d) In the RNA polymerase I (pol I) pre-initiation complex (PIC), there are a multitude of protein–protein and protein–ribosomal promoter–DNA interactions (i) –(vi). For simplicity, only a single dimer of UBF contacting the rDNA is shown, but there might be a dimer at both the UCE and core sequences. UBF binds pol I, in part, through Pol-I-subunit PAF53, a homologue of yeast pol-I-subunit A49. Also involved in PIC formation, but of unknown identity, are TIF-IC, a mouse Pol-I-associated factor [41], and p70 [81]. The polymerase core subunits and pol-I-associated factors, other than hRRN3, are not detailed.