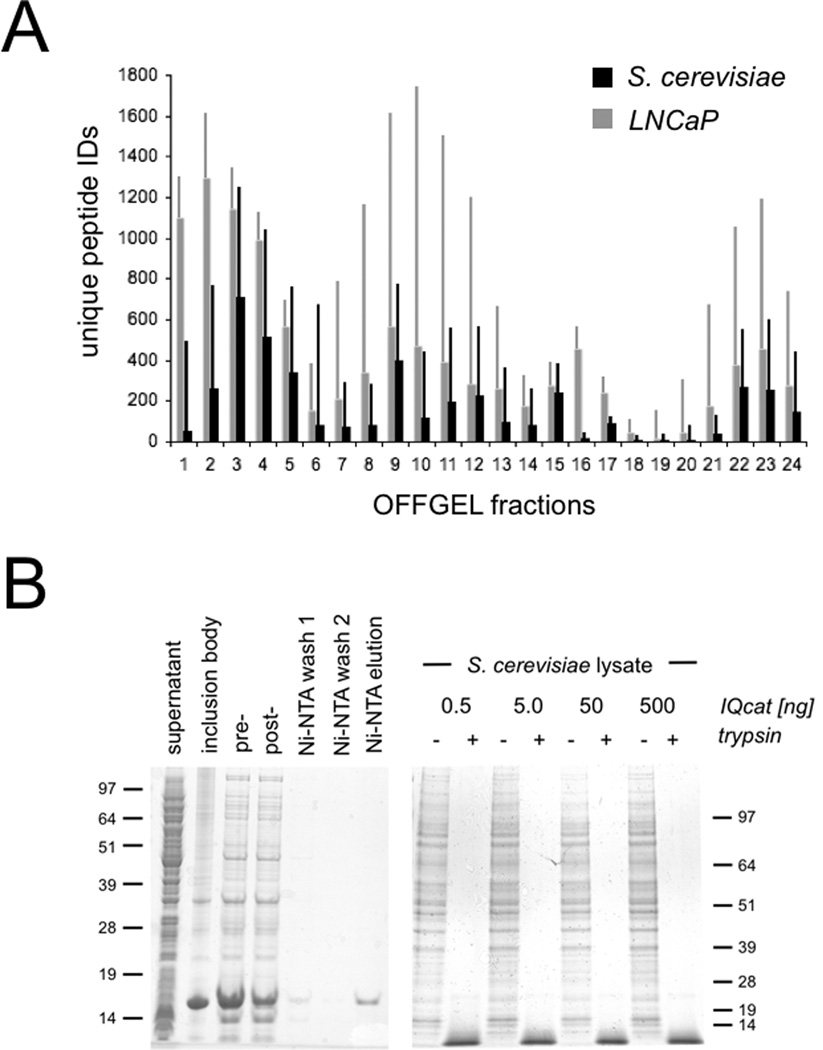
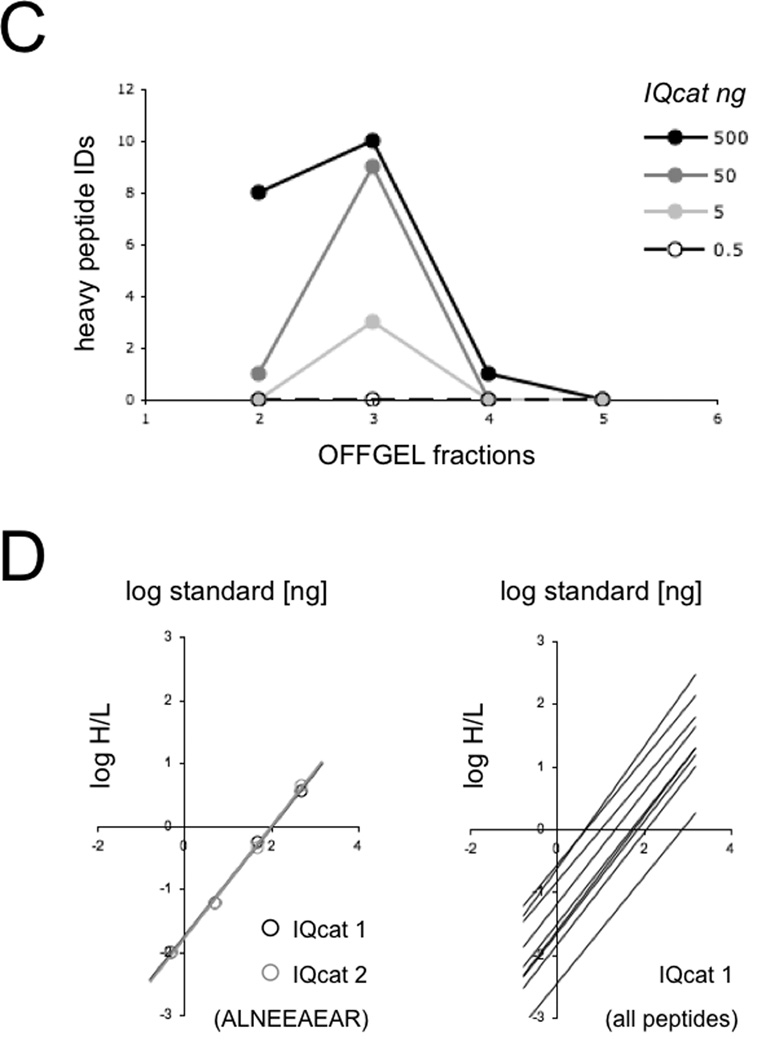
Figure 1.
IQcat Design and Measurement A) Total peptide IDs (thin bars) and peptide IDs unique to an individual OFFGEL fraction (thick bars) were determined from shotgun analysis of LNCaP and S. cerevisiae lysates. B) Isotopically labeled IQcat was purified from inclusion bodies by Ni-NTA (left), and co-digested in spike-in samples with 0.5, 5, 50, or 500 ng heavy IQcat to 25 µg S. cerevisiae lysate (right). Proteins are visualized by SDS-PAGE Coomassie stain of 12% and 4–12% gradient gels. C) OFFGEL fractionation of spike-in samples measured by LC-MS as the number of unique heavy IQcat peptide identifications per fraction. D) Log/Log plots of peptide heavy to light ratios give linear and overlapping correlations for the ALNEEAEAR peptide in both IQcat constructs (left). Linear fits for IQcat 1 peptides show a range of endogenous peptide concentration spanning two-log orders.