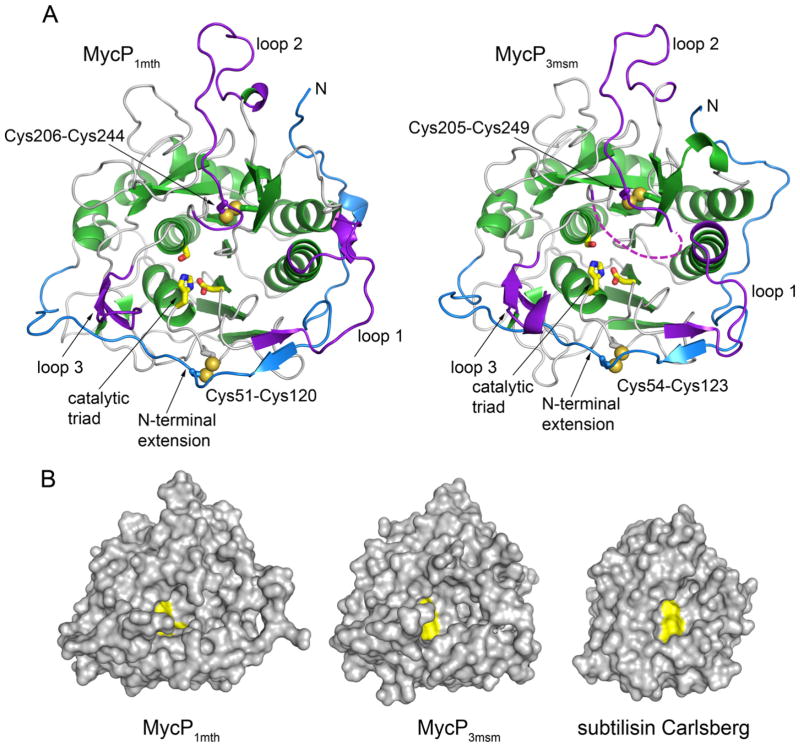
Fig. 2. Crystal structures of MycP1mth and MycP3msm.
A. The structures are shown in the same orientation with α helices and β strands of core subtilisin domain in green, the insertion loops in purple and the N-terminal extension in blue. The catalytic triad residues (yellow) are shown as sticks. Bound ions are omitted for clarity. The disordered amino acid residues in loop 2 of MycP3msm are indicated by a purple dashed line.
B. Structures of MycP1mth and MycP3msm and subtilisin Carlsberg (PDB 3UNX) (Fisher et al., 2012) are shown in surface representation with catalytic triad residues highlighted in yellow.