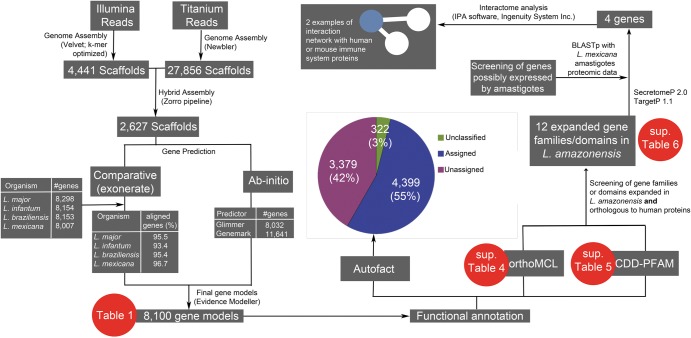
Figure 2.
Bioinformatics analysis workflow used in the present study. Sequenced reads from the L. (L.) amazonensis genome were assembled into 2627 scaffolds and 8100 genes were predicted using comparative and ab initio prediction tools. The functional analysis of these predicted genes included: (i) AutoFACT functional annotation, which revealed that 45% of the predicted genes were unclassified or with unassigned function; (ii) screening for orthologous families of genes among Leishmania spp. (OrthoMCL); and (iii) screening for information about conserved protein domains deposited in CDD and PFAM databases. Expanded or exclusive orthologous proteins, or those conserved domains detected in the L. (L.) amazonensis genome were selected for interactome analysis with mammalian host proteins. This selection involved screening for possibly secreted proteins (using SecretomeP and TargetP) that also were orthologous to immune function-related proteins in humans and mice.