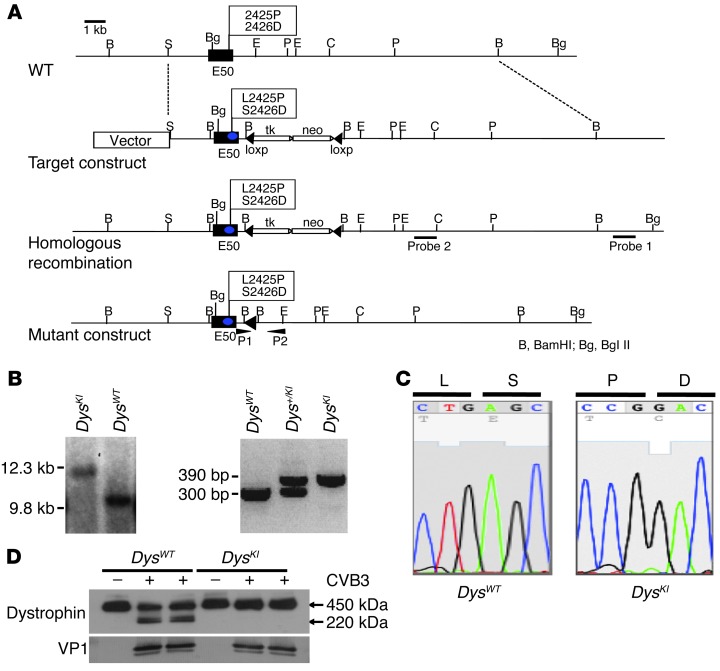
Figure 1. Generation of enteroviral protease 2A cleavage–resistant DysKI mice.
(A) Genomic structure and design of DysKI mouse. We changed the wild-type exon 50 (E50) amino acid residues 2425 and 2426 from leucine to proline (L2425P) and serine to aspartic acid (S2426D), respectively. (B) Confirmation of DysKI recombination by Southern blotting of mouse embryonic stem cells. Genomic DNA isolated from embryonic stem cells was used to confirm that homologous recombination had occurred as opposed to random insertion. Southern blotting with probe 1 was performed with a BglII digest, demonstrating loss of the wild-type genome at 9.8 kb (left panel). Female mice were genotyped using PCR primers P1 and P2 (right panel). Data are shown for mice with wild-type dystrophin (DysWT), heterozygous dystrophin knockin (Dys+/KI), and homozygous dystrophin knockin (DysKI). (C) Sequence analysis of exon 50 showing replacement of nucleotides in DysKI mouse genomic DNA as expected. (D) Western blot showing dystrophin in embryonic cardiac myocytes 24 hours after CVB3 infection, in which the cells were isolated from DysWT and DysKI mice. Dystrophin cleavage was inhibited in myocytes from DysKI mice. Comparable levels of viral infection of the cells are demonstrated by the presence of the CVB3 capsid protein VP1.