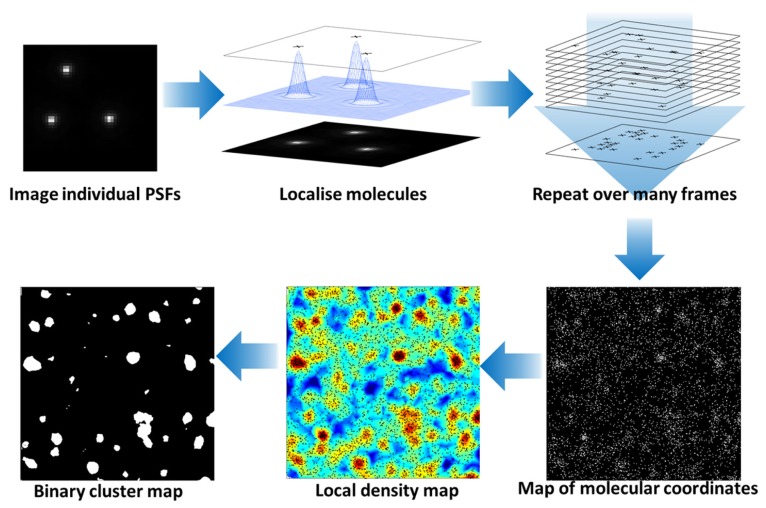
FIGURE 1.
Photoactivated localization microscopy analysis of protein clustering at the cell surface. Photo-activation or stable dark states of fluorophores are exploited to limit the number of fluorescent molecules in each image frame. The fluorescence of individual molecules are captured with a camera and the center of the point-spread function (PSF) calculated to localize the molecules with nanometer precision. During the imaging processes, the fluorescence molecules are bleached so that the combination of photo-activation and photo-bleaching gives the appearance that molecules “blink” during the acquistion. Over successive frames, an image of all fluorescent molecular positions is built up. The molecular coordinates can be used to generate an image and be quantitatively analyzed to reveal the local density of fluorescent molecules (here based on Ripley K-function before and after application of a threshold) and hence clusters of proteins at the plasma membrane identified. For details on the cluster analysis, please see Williamson et al. (2011).