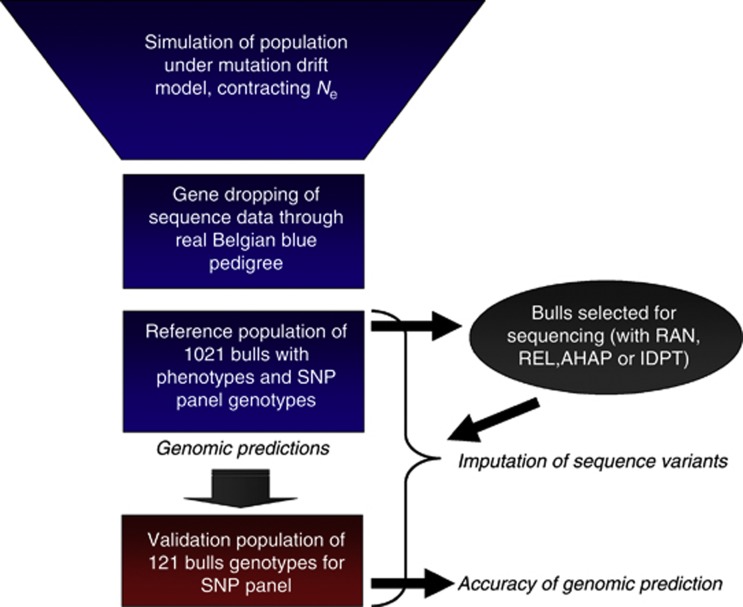
Figure 1.
Simulation scheme. A base population was simulated according to the inferred demography of Bos Taurus cattle of Macleod et al. (2012A). The demography has a large ancient effective population size contracting to a much smaller effective size following domestication and breed formation. Fregene was used to simulate 50 MB of sequence under a mutation drift model with this demography. From this population, the founders of the real Belgian Blue cattle breed-recorded pedigree were selected at random, and the sequence data were gene-dropped through the entire pedigree with recombination. A subset of bulls was selected from the pedigree, on the basis that they had DNA samples available. These bulls were ‘genotyped' for a dense SNP panel. A much smaller subset of the bulls were selected for sequencing, with an Illumina sequencing process simulated. The sequence data was then imputed into all bulls with SNP panel genotypes. A prediction equation, based either on SNP panel genotypes or imputed sequence data, was estimated in the reference population. Finally, accuracies of genomic prediction were assessed in the validation population.