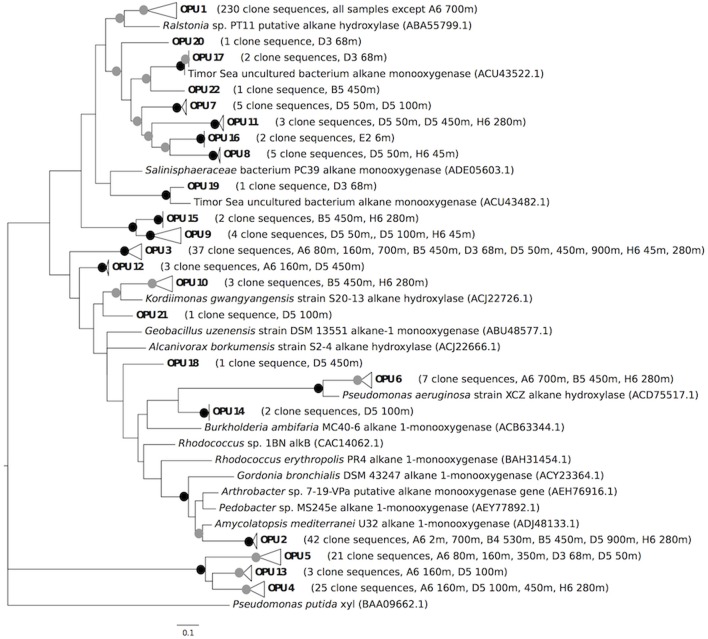
Figure 1.
Maximum likelihood tree comparing putative alkB amino acid sequences from this study with reference alkB sequences obtained from other studies. Bootstrap values from 100 resamplings are indicated with black circles for values of 95–100% and gray circles for values of 50–94%. OPUs were determined using a distance cutoff of 0.20 (80% sequence similarity). OPU 1 clusters with the following reference sequences: Alcanivorax borkumensis SK2 alkane 1-monooxygenase (CAL18155.1), Alcanivorax borkumensis S12-4 alkane hydroxylase (ACJ22702.1), Alcanivorax dieselolei S10-17 alkane hydroxylase (ACJ22698.1), Marinobacter aquaeolei VT8 alkane 1-monooxygenase (ABM17541.1), and Marinobacter sp. S17-4 putative alkane monooxygenase (ACT31523.1). OPU 3 clusters with Marinobacter adhaerens HP15 alkane 1-monooxygenase (ADP98338.1), Marinobacter hydrocarbonoclasticus S17-4 alkane hydroxylase (ACJ22716.1), and Marinobacter sp. P1-14D alkane hydroxylase (ACS91348.1). The tree was rooted with a xylene monooxygenase amino acid sequence from Pseudomonas putida (Hara et al., 2004).