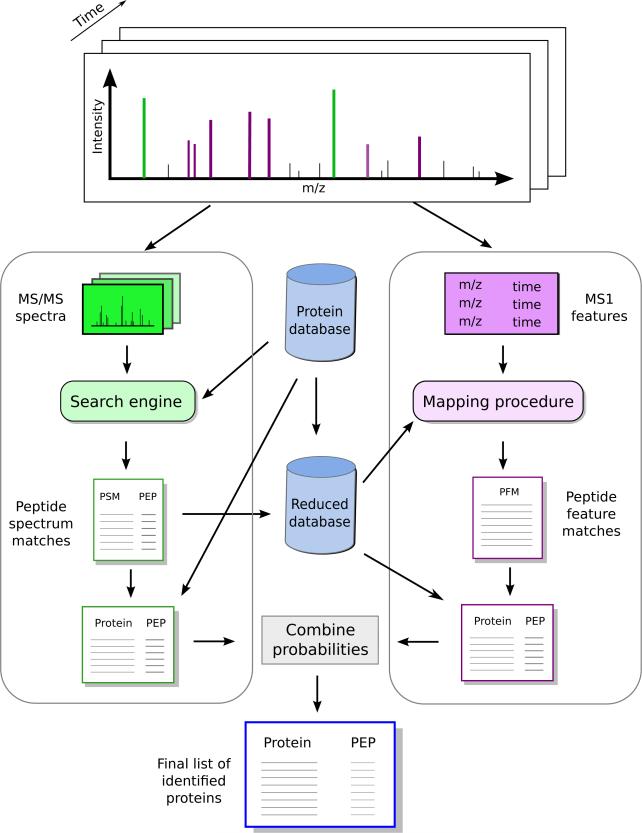
Figure 3. The in silico AMT workflow.
Our approach includes two complementary workflows. The first one, depicted on the left-side, includes the typical steps of a shotgun experiment, where we employ the fragmentation spectra to derive protein probabilities. In a second workflow, we map the peptides that did not match a fragmentation spectrum to the MS1-features, and use the resultant peptide-feature matches to derive a new set of protein probabilities. Finally, we combine the two statistics to derive improved protein-level confidence measures.