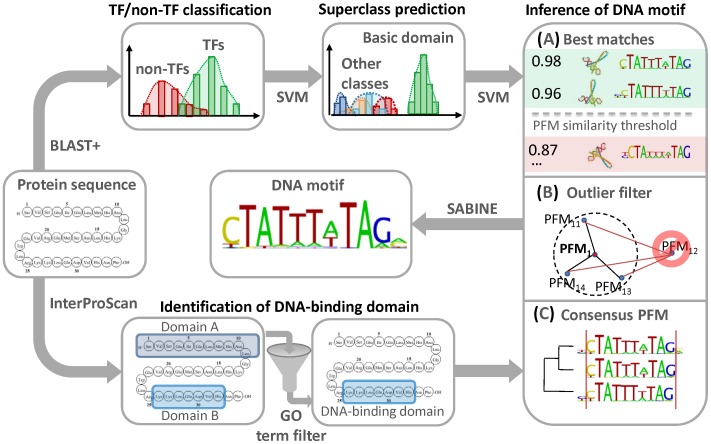
Figure 1. Bioinformatics pipeline for the structural and functional annotation of transcription factors.
First the input protein sequence is aligned to a non-redundant protein database using the BLAST heuristic. The bit score distributions of the TFs and non-TFs among the BLAST hits are represented by means of percentiles. These percentiles are incorporated into SVM classifiers for the discrimination of TFs from non-TFs (Step 1). If a given protein sequence was classified as a TF, another SVM is applied to predict its structural superclass (Step 2). The tool InterProScan is used to predict the functional domains of the TF and the DNA-binding domains among these are identified based on the associated GO terms (Step 3). Finally, the tool SABINE infers a DNA motif using an SVR-based algorithm (see Methods section) that takes the structural superclass and DNA-binding domains of the TF as input (Step 4).