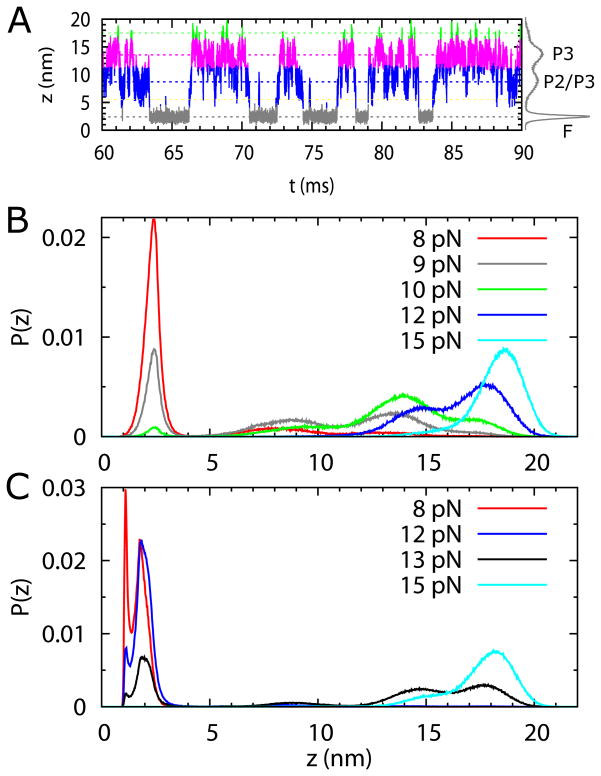
Figure 5.
(A) Time trace of extension, z, of the SAM-III riboswitch without the metabolite at f = 9 pN. (B) and (C) From time traces, such as the one in (A), we calculated the probability distributions, P(z)s of extension z. The time traces were obtained from constant force simulations with each at least 150 ms long. (B) Distributions P(z)s in the apo state of the RNA at various f values. P(z) at f = 9 pN (green curve) shows four peaks, corresponding to F, P2/P3, and P3 states, and one at z ~ 17.5 nm representing the unstable U state. The ΔP1 state (P1 unfolded, with other helices intact), at z ~ 5.5 nm, is visited transiently, with very low probability. The riboswitch stays mostly unfolded when f > 11 pN. In the force range, 9 pN < f < 11 pN, the P3 folded state is the most stable. At f = pN the riboswitch iss fully folded (red curve). The riboswitch goess from the unfolded to the folded state following the pathway, U → P3 → P2/P3 → F, when force decreases from a large to a small value. (C) Same as (B) except SAM is bound to RNA showing that the intermediate states in (B) are only transiently present. In the presence of SAM, the riboswitch stays mostly unfolded when f > 13 pN. As f decreases to below 13 pN, the riboswitch transits to the folded state with z = 2 nm with G1 and U53 not paired (Fig. 1A). At forces f < 8 pN, G1 and U53 forms a base pair with z = 1 nm.