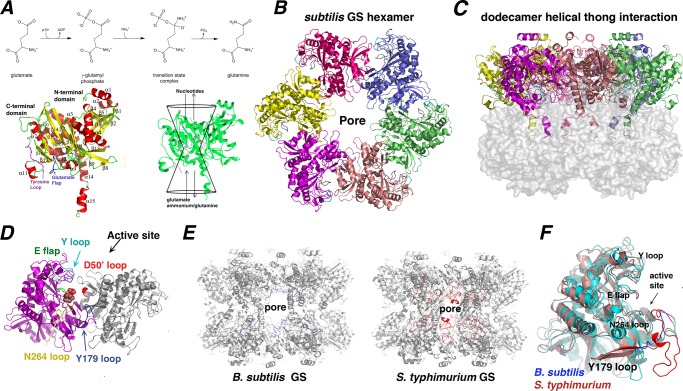
FIGURE 1.
Structure and enzymatic mechanism of B. subtilis GSI-α. A, top, a scheme depicting the GS reaction mechanism. Bottom left, secondary structural elements of the GSI-α subunit. For reference, the Glu flap is colored blue, and the Tyr loop is colored magenta. Ribbon diagrams figures were made using PyMOL (47). Bottom right, the GS active site is illustrated as a bifunnel with entrance and exit portals. The glutamate and ATP substrates enter and bind at opposite ends of the bifunnel, and the respective products leave via the same portals. B, a hexamer of the dodecameric B. subtilis GS viewed down the molecular 6-fold axis. C, the GS dodecamer rotated by 90º relative to Fig. 1B. In this figure, the “bottom” hexamer is shown as a surface to highlight the interhexamer contacting α14-α15 helices. D, close-up view of an active site created by neighboring subunits. Key active site loops that line the active site are labeled and colored differently. A bound glutamate is shown in CPK. E, comparison of GS dodecamers of the B. subtilis GSI-α and S. typhimurium GSI-β proteins. Shown in different colors (B. subtilis in blue and S. typhimurium in red) are the active site Tyr179 loops that line the dodecameric pore (labeled). F, superimposition of a B. subtilis subunit (blue) onto a S. typhimurium subunit (red), highlighting their strong structural correspondence except for the Tyr179 loop. The location of the pore is labeled in B and E for reference.