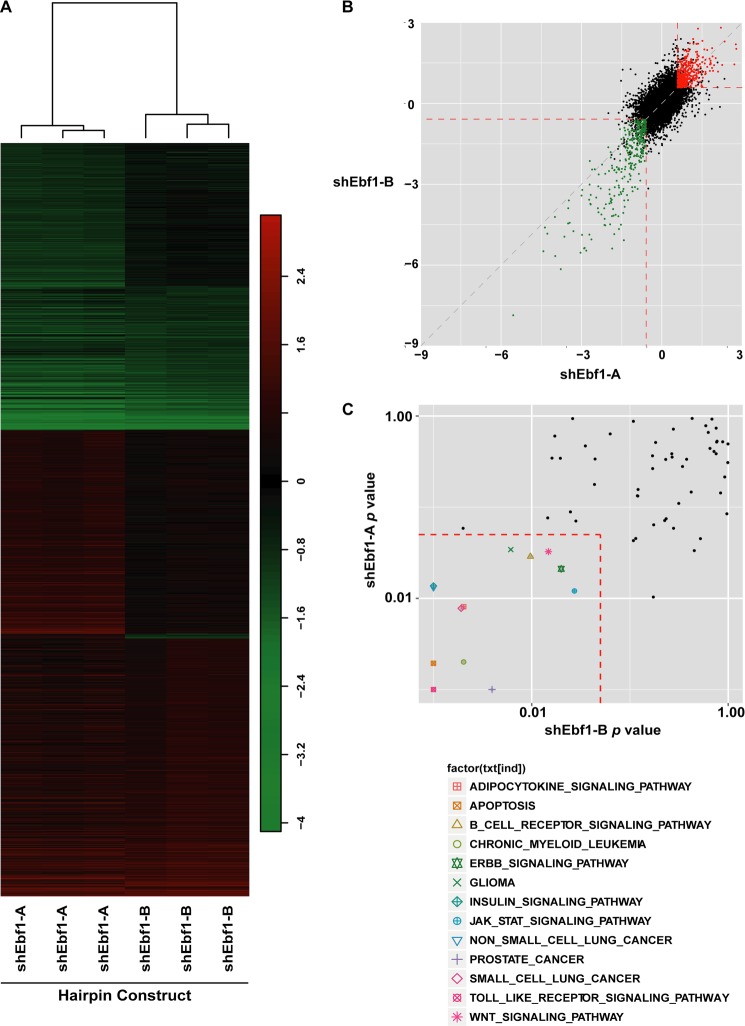
FIGURE 2.
Microarray and gene set enrichment analysis of EBF1-deficient adipocytes. A, heat map of probes whose signals were significantly up- or down-regulated (p < 0.01 and -fold change of >1.5 or <1/1.5) in cells infected with lentivirus harboring either of two shEbf1 constructs. Clustering of the two hairpins is indicated by the dendrogram. y axis, probes; x axis, sample identity. Vertical scale indicates log(-fold change). B, scatter plot comparing the relative expression of individual genes (shown as dots) in cells treated with shEbf1-A versus shEbf1-B. Genes significantly (p < 0.01) down-regulated by both hairpins are shown in green, and genes significantly (p < 0.01) up-regulated by both hairpins are shown in red. The axis indicates log(-fold change) with each hairpin. C, scatter plot comparing the p values of enriched KEGG pathways in cells treated with shEbf1-A versus shEbf1-B. KEGG pathways that scored significantly (p < 0.05) with both shEbf1 constructs are shown inside the dotted box.