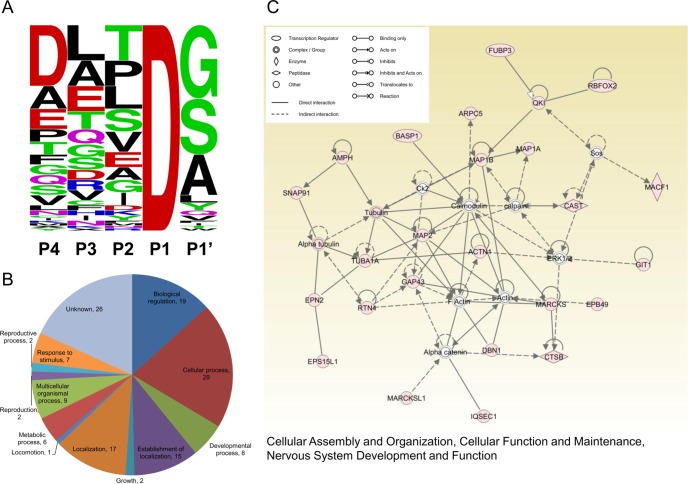
Fig. 2.
Motif composition, Gene Ontology analysis and Ingenuity Pathway Analysis of putative caspase substrates. A, Sequence-logo plot representing amino acid frequency in P4 to P1′ at 81 unique putative caspase cleavage sites identified in STS-treated neurons (generated by WebLogo, http://weblogo.berkeley.edu/logo.cgi). B, Biological Process Gene Ontology (GO) terms annotating the 56 putative caspase substrates identified in STS-treated neurons (NCBI annotations, analyzed by Scaffold). The numbers of substrates allocated to each GO term are indicated. It should be noted that one substrate could be annotated by multiple GO terms. C, The highest-scored network (score 63, focus molecular 25) identified by Ingenuity Pathway analysis of the 56 putative substrates. Colored nodes indicate putative substrates used in the pathway analysis.