Table 2.
The most potent inhibitors of RNA editing and their possible mode of action.
| ID/source | Structure | IC50 (μM) based on adenylylation activity | IC50 (μM) based on full-round RNA editing activity | Potential mode of inhibition | References |
|---|---|---|---|---|---|
| V4, Sigma Mordant Black 25 | 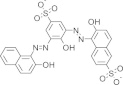 |
1.59 ± 1.10 | N.D. | Adenylylation | Durrant et al. (2010) |
| C35/V2, NSC162535 | 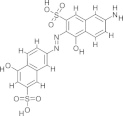 |
1.53 ± 1.17 | N.D. | Deadenylylation/adenylylation/RNA–protein interaction | Durrant et al. (2010), Moshiri et al. (2011) |
| V1, NSC45609 | 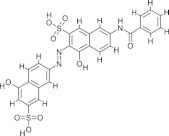 |
2.16 ± 1.20 | N.D. | Adenylylation | Durrant et al. (2010) |
| V3, NSC1698 | 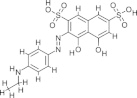 |
8.36 ± 1.71 | N.D. | Adenylylation | Durrant et al. (2010) |
| S5, NSC16209 | 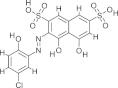 |
1.01 ± 0.16 | 2.1 ± 0.4 | Deadenylylation/adenylylation/RNA–protein interaction | Amaro et al. (2008), Moshiri et al. (2011), Moshiri and Salavati (2010) |
| S1, NSC100234 | 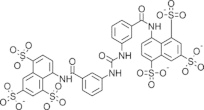 |
1.95 ± 0.61 | N.D. | Adenylylation | Amaro et al. (2008) |
| NF023 | 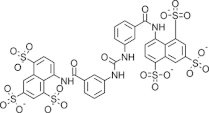 |
N.D. | 2.9 ± 0.7 | RNA editing before or at the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) |
| Mitoxantrone | 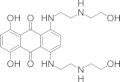 |
N.D. | 2.6 ± 0.5 | RNA editing before or at the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) |
| D-Sphingosine | N.D. | 2.0 ± 0.1 | RNA editing after the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) | |
| GW5074 | 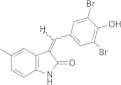 |
N.D. | 2.9 ± 0.4 | RNA editing before or at the endonuclease cleavage step | Liang and Connell (2010) |
| Protoporphyrin IX | 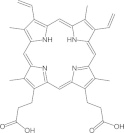 |
N.D. | 0.9 ± 0.1 | RNA editing before or at the endonuclease cleavage step | Liang and Connell (2010) |
N.D.: not determined.
