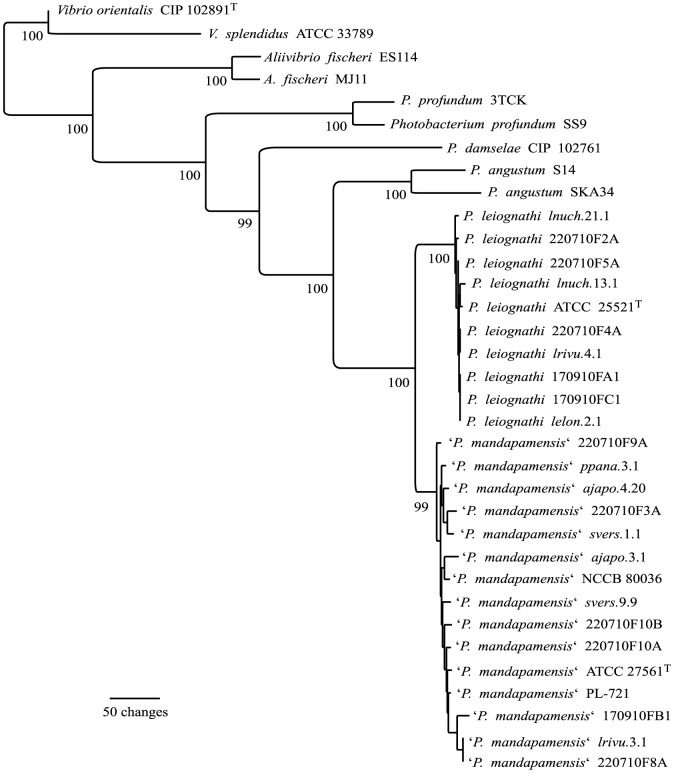
Figure 1. Phylogenetic analysis of P. leiognathi and ‘P. mandapamensis’.
Phylogenetic tree representing 1 of 45 equally parsimonious hypotheses resulting from the analysis of concatenated alignments of nucleotide sequences of PMSV_2285 and PMSV_ 4043 homologs in 25 P. leiognathi and ‘P. mandapamensis’ strains and related Vibrionaceae. Sequence alignment had 1794 characters (including 1020 informative characters). The trees length was equal to 3030. The 45 resulting hypotheses differed in the predicted relationship between strains within P. leiognathi and ‘P. mandapamensis’ clades. Jackknife resampling values are shown at the nodes, some were omitted for clarity. Analyses of the concatenated PMSV_2285 and PMSV_ 4043 alignment were also carried out using neighbor-joining and maximum-likelihood algorithms; results of these analyses showed the same cladding of P. leiognathi and ‘P. mandapamensis’ strains (see Figure S2).