Abstract
The N-oxygenation of an amine group is one of the steps in the biosynthesis of the antibiotic chloramphenicol. The non-heme di-iron enzyme CmlI was identified as the enzyme catalyzing this reaction through bioinformatics studies and reconstitution of enzymatic activity. In vitro reconstitution was achieved using phenazine methosulfate and NADH as electron mediators, while in vivo activity was demonstrated in Escherichia coli using two substrates. Kinetic analysis showed a biphasic behavior of the enzyme. Oxidized hydroxylamine and nitroso compounds in the reaction were detected both in vitro and in vivo based on LC–MS. The active site metal was confirmed to be iron based on a ferrozine assay. These findings provide new insights into the biosynthesis of chloramphenicol and could lead to further development of CmlI as a useful biocatalyst.
Keywords: Chloramphenicol biosynthesis, Arylamine oxygenation, N-Oxygenation, Di-iron oxygenase
1. Introduction
Aromatic nitro groups are relatively rare functional groups in natural products, but are found in diverse types of important antibiotics, such as chloramphenicol, pyrrolnitrin, aureothin, spectinabilin, azomycin, and rufomycin.1,2 The biosynthesis of aromatic nitro groups in natural products is poorly understood, and in particular the enzymes involved in catalyzing the formation of aromatic nitro groups are not well characterized. To date, only two N-oxygenases, PrnD3–5 and AurF6–10 that are involved in the biosynthesis of pyrrolnitrin and aureothin, respectively, have been extensively characterized.
Chloramphenicol is a potent broad-spectrum antibacterial antibiotic produced by Streptomyces venezuelae and certain other actinomycetes.11 Because of its medical importance and unusual chemical structure (i.e., aromatic nitro group and dichloroacetamide moiety), the biosynthesis of chloramphenicol has been extensively investigated for more than four decades, mostly by Vining et al.11–15 The gene cluster for chloramphenicol biosynthesis has been identified and cloned, with expression in a heterologous host; however, the cluster was not fully characterized.14,16 In particular, the enzyme that catalyzes the formation of a nitro group in chloramphenicol remains unclear. Vining et al. proposed the final step of chloramphenicol biosynthesis to be the arylamine oxidation, based on feeding experiments with isotopically labeled compound 1.12 However, no biochemical or genetic evidence has been provided to support this hypothesis. Thomas et al. suggested that the arylamine intermediate might be oxidized by CmlI while the substrate is still bound to CmlP, a non-ribosomal peptide synthase.17 This is not without precedent; Lipscomb et al. recently characterized CmlA, which hydroxylates the l-p-amino-phenylalanine as it is still attached to CmlP.18 To shed light into this issue, we sought to use a variety of methods, such as bioinformatics, DNA cloning, and both in vitro and in vivo reconstitution of the enzyme to investigate the functional role of CmlI in chloramphenicol biosynthesis.
2. Results and discussion
Bioinformatic analysis indicated that CmlI shares ~ 34% amino acid sequence identity with AurF. More importantly, like AurF, two copies of the EX28–37DEXXH motifs are present in this protein. This motif is also conserved in several other di-iron oxygenases.7,19,20 Based on these results, we hypothesized that CmlI is a non-heme di-iron N-oxygenase. Subsequently, cmlI from the genomic DNA of S. venezuelae (NRRL ISP-5230) was cloned into pET26 vector with a His6-tag. DNA sequencing data suggested that there were a few errors in the previously reported sequence of cmlI. As a result, the total number of amino acids in our CmlI is 339 rather than the previously reported 338.14 The former number should be correct because the more recently sequenced genome S. venezuelae ATCC 10712 also gives 339 amino acids for CmlI. This inevitably changes the translated sequence to a small degree, albeit it does not affect the conserved EX28–37DEXXH motifs (Fig. S1).
To study the activity in vitro, CmlI with an N-terminal His6-tag was over-expressed in Escherichia coli and further purified by affinity chromatography to be >95% pure (Fig. S2). Upon concentration, the enzyme appeared light yellow in solution, suggesting the existence of metals or other cofactors.
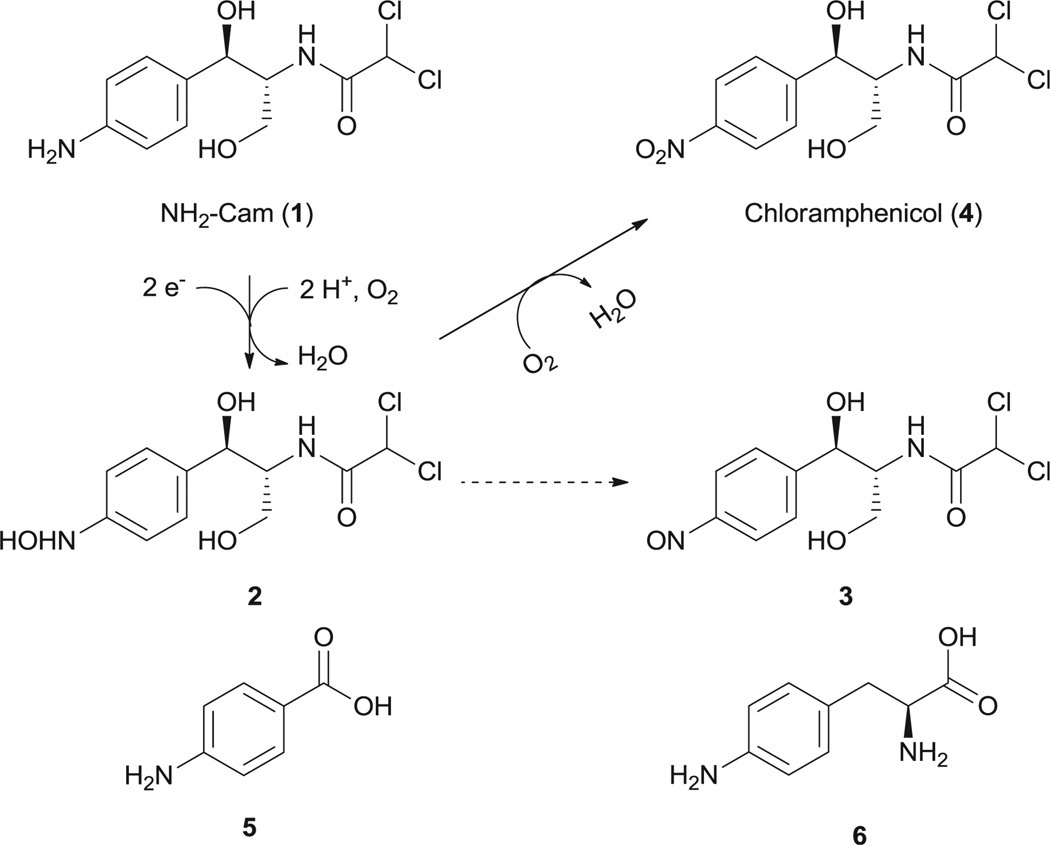
The initial synthesis of substrate 1 (1R,2R)-(−)-2-(N-dichloroacetyl)-amino-1-(4-aminophenyl)-1,3-propanediol (Scheme 1, termed NH2–Cam hereafter) following the reported catalytic hydrogenation procedure only resulted in de-chlorination of chloramphenicol.21 The synthesis was finally achieved by canonical acylation of a NH2–Cam precursor with dichloroacetyl chloride (Supplementary data).
Scheme 1.
Compounds studied in this paper. The mechanism for conversion of NH2–Cam (1) to chloramphenicol (4) by CmlI is given above. Solid lines indicate reactions catalyzed by CmlI, and dashed lines indicate a spontaneous nonenzymatic oxidation.
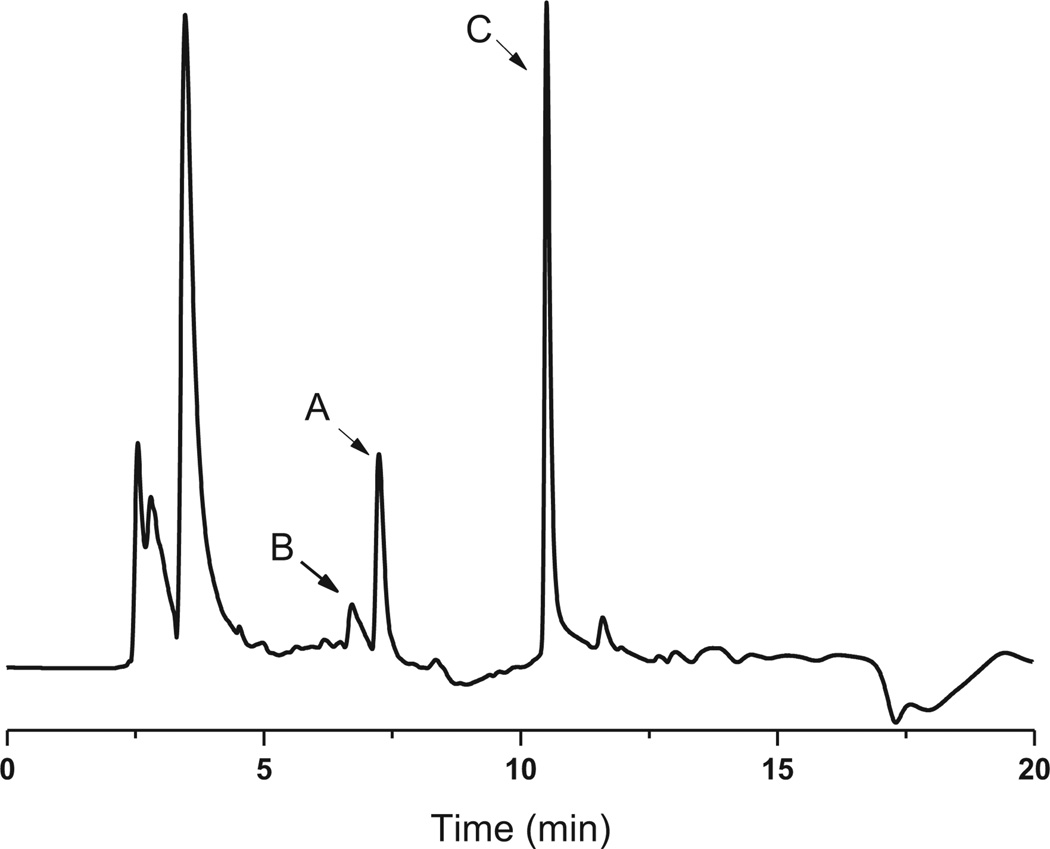
In the initial enzymatic assay, no conversion of NH2–Cam was observed when mixed with purified CmlI. Inspired by our previous studies on AurF, we envisioned that the in vitro oxidation of NH2–Cam by CmlI might require proper electron mediators.7 These mediators serve to replace the native enzymatic electron transfer systems such as ferredoxins. Three such compounds, namely phenazine methosulfate (PMS), phenazine ethosulfate (PES), and flavin mononucleotide (FMN), were tested in the presence of NADH, NH2–Cam, and CmlI. LC–MS analysis of the resulting mixtures confirmed our hypothesis. The conversion of NH2–Cam to Cam by CmlI was observed in the presence of PMS (Fig. 1) or PES (data not shown). We attempted to determine the kinetic parameters of CmlI. However, the reaction showed a biphasic behavior, with a fast step consuming significant amounts of NH2–Cam within 15 s, and a slower step afterward (Fig. S3). Similar behavior was seen when production of 4 was monitored. Addition of different starting amounts of enzyme showed corresponding increases in rate for both steps with increasing enzyme concentration. Essentially complete depletion and conversion of the substrate to product was observed within 30 min using 15 µM of enzyme. Adding more enzyme to the reaction during the slower step caused no significant increase in activity for NH2–Cam depletion but a short burst of chloramphenicol production (data not shown).We did not observe this biphasic behavior in AurF using similar conditions. Regardless, these data indicates that CmlI is a bona fide N-oxygenase involved in chloramphenicol biosynthesis.
Fig. 1.
HPLC trace of CmlI-catalyzed chloramphenicol formation in vitro at 285 nm. (A) Starting material NH2–Cam (1); (B) hydroxylamine intermediate (2); (C) nitroso compound (3) and final product chloramphenicol (4).
The oxidative steps in converting arylamine to arylnitro compounds have been discussed for AurF and PrnD, where aryl-amine was first oxidized to the hydroxylamines and nitroso compounds.3,6 Our effort to detect these intermediates in the reaction mixture of CmlI with NH2–Cam enabled us to identify these compounds. As shown in Fig. 1, the hydroxylamine intermediate (B) elutes before the NH2–Cam (A), while the nitroso side product showed up with a retention time very close to or overlapping with chloramphenicol in all HPLC conditions tested (C). Intermediates were confirmed in negative mode LC–MS by the characteristic Cl isotope pattern and detection of the correct parent ions (Fig. S4). Recent work by Bollinger et al. has shown that for AurF, the aryl hydroxylamine can be converted to the nitro compound without input of endogenous electrons, and we speculate that a similar mechanism may be at play in CmlI.23,24We attempted to synthesize 2 via Sn(II) mediated reduction of 4; however its oxidation to 3 occurs spontaneously non-enzymatically and we could not obtain pure enough compound for 2. We were able to synthesize 3 according to the procedures in literature.22 As 3 could be a dead-end or non-native intermediate, we speculated that the biphasic behavior observed may be due to competitive inhibition by the nitroso compound. However, upon addition of varying amounts of 3 to reactions containing 10 µM of CmlI, there is only a slight decrease in the initial burst phase, and no real change in the rate of the second slow phase (Fig. S5).Thus, it is unlikely that the nitroso compound is responsible for the biphasic behavior of the enzyme, and some other unknown mechanism is in fact responsible for the reaction profile.
In vivo reconstitution of CmlI was also attempted. Addition of compound NH2–Cam to E. coli expressing cmlI gave larger depletion of the substrate compared to controls, and appearance of peaks corresponding to the hydroxylamine, nitroso, and nitro compounds (Fig. S6). This was confirmed by detection of the product on HPLC, and detection of the correct masses on LC–MS for the intermediates. Similarly, feeding of p-aminobenzoic acid (5) showed some conversion of the substrate to the hydroxylamine, but the production of the nitro compound was not conclusively detected. The conversion of l-4-aminophenylalanine (6), an intermediate in chloramphenicol biosynthesis, could not be determined, as the substrate was consumed at similar rates in both the sample and control, and the nitro product was not detected. The activity of CmlI on free substrates 1 and 5 suggests, but does not prove, that the N-oxidation occurs while the native substrate is not tethered to the native NRPS CmlP.16,17 It has been noted that the AurF can be reconstituted in the heterologous host due to a ribonucleotide reductase-like electron transfer pathway, and CmlI likely works by a similar mechanism by using reduced ferredoxins as a source for electrons.8 This is supported by the strict conservation of residues involved in electron transfer for CmlI and AurF (Fig. S1). Residues W44, D143, and H239 in CmlI correspond to side chains used in AurF’s electron transfer (W35, D135, and H230). However residues involved in ferredoxin binding are not strictly conserved. As CmlI is also able to turn over the much smaller substrate 5, the presence of a keto or a hydroxyl group at the benzylic position may be sufficient for recognition as a substrate for at least the initial oxidation. This is similar to what has been found for AurF, but as CmlI can accept much larger substrates, it may be more amenable for engineering as an N-oxidation catalyst.8 It remains to be seen what other factors determine the substrate specificity of CmlI.
Various metals have been found in oxygenases to mediate substrate oxidation.25–28 Although it is clear now that AurF, a homolog of CmlI, is a non-heme di-iron N-oxygenase, we cannot rule out the existence of other metals in CmlI. To address this issue, we analyzed by ICP-MS the metal contents in CmlI purified from cells cultivated in Luria–Bertani medium. Only Fe was found to be significantly present. To rule out the possible involvement of Mn in catalysis, which was once suggested for AurF,10 the ratio of Mn and Fe in CmlI were further assessed for CmlI purified cells cultivated in either Luria–Bertani medium or M9 minimal medium with equal amounts of Fe and Mn added. The concentration of Fe in the protein was much higher than that of Mn, by about 17-fold in both cases. The Fe/CmlI ratio ranged from 1.2 to 1.4 (Table 1). When we used the iron-specific ferrozine assay,29 the Fe to CmlI ratio was found to be 2:1 (Table 1). Taken together, our data seem to suggest that CmlI, just like AurF, is also a di-iron oxygenase.
Table 1.
Metal analysis of CmlI
| Fe/Mn ratio (ICP) |
Fe/CmlI ratio (ICP) |
Fe/CmlI ratio (ferrozine) |
|
|---|---|---|---|
| CmlI from M9 | 17 | 1.4 | 2.2 |
| CmlI from LB | 17 | 1.2 | 1.8 |
3. Conclusions
We have reported the first biochemical characterization of the N-oxygenase CmlI and confirmed its role in chloramphenicol biosynthesis. Reconstitution of the enzyme activity was achieved in vitro via addition of chemical reductants and in vivo in E. coli. Furthermore, our data show that CmlI, just like AurF, is a non-heme di-iron oxygenase. These findings have opened the door for further mechanistic and structural studies of this type of oxygenases and biocatalyst development using protein engineering tools.
4. Materials and methods
4.1. Bacteria strains and culture conditions
The wild-type strain of S. venezuelae ATCC 10712 was obtained from the American Type Culture Collection (ATCC, Manassas, VA). S. venezuelae and its derivatives were maintained on ISP2 agar medium and cultivated in YGM liquid medium (malt extract 5 g/L, yeast extract 2 g/L, glucose 2 g/L) at 28 °C with shaking at 250 rpm. Genomic DNA was isolated using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI) using standard protocols.
4.2. Overexpression and purification of CmlI
The cmlI gene was amplified by PCR from the genomic DNA of S. venezuelae (NRRL ISP-5230) and cloned into pET26 vectors (Novagen, Gibbstown, NJ) using a forward primer (5′-CAGTTCATATGCGTGACCACACGGACGAGAAATC-3′) and a reverse primer (5′-GTAGTCAAGCTTTCATCGGG TCACCGTCGTGC-3′) between NdeI and HindIII sites (underlined sequences). The corresponding plasmid was transformed into E. coli BL21 (DE3) cells. Kanamycin (50 µg/mL) was used as a selection marker. For expression of cmlI in Luria–Bertani medium, cells were grown at 37 °C to OD600=0.6–0.8. Then protein expression was induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the cells were agitated at 250 rpm and 25 °C overnight. For expression of CmlI in M9 medium, cells were grown in M9 medium at 37 °C to OD600=0.6–0.8. To the culture were added l-methionine (60 mg/L), l-leucine (50 mg/L), l-isoleucine (50 mg/L), l-valine (50 mg/L), l-lysine (100 mg/L), l-threonine (100 mg/L), l-phenylalanine (100 mg/L), (NH4)2Fe(SO4)2 (0.5 mM), and MnSO4 (0.5 mM). Protein expression was induced by IPTG (0.1 mM), and cells were agitated at 250 rpm and 18 °C overnight. The cell lysate was prepared by passing cells resuspended in buffer (20 mM Tris–HCl, pH=7.5, 100 mM NaCl, 1 mM β-mercaptoethanol, 5% glycerol) twice through French Press®. N-His6-tagged CmlI was purified off Co-column to over 95% purity. Purified CmlI was then dialyzed against storage buffer (50 mM HEPES, 10% glycerol). Protein was quantified using the Bradford Quickstart Reagent (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard.
4.3. In vitro and in vivo assays
In vitro assays of CmlI were performed on a 600 µL scale. The final reaction mixture contained CmlI (5–20 µM) substrate NH2–Cam (250 µM), PMS (100 µM), NADH (1 mM), and HEPES (20 mM, pH=7.5). Reactions were performed at room temperature (23–25 °C) and were started by addition of NADH to the reaction mixture. Reaction was quenched by trifluoroacetic acid (final concentration 0.8% v/v) and centrifuged. For in vivo reconstitution of CmlI, cells were grown similar to cells used for purification, except that Terrific Broth medium was used instead of Luria–Bertani broth. Cells were pelleted by centrifugation, washed twice in assay buffer (25 mM MOPS, 8 g/L NaCl, 1 g/L KCl, 2 g/L glucose, pH 7.2), and resuspended to a final OD600 of 15 in a final volume of 50 mL. Substrate was added to a final concentration of 250 µM, and the reaction was followed for 3 h at 30 °C, taking time points every 30 min. As a control, cells containing only the vector were used, and subjected to the same analysis and time course.
4.4. LC–ESI-MS and ESI-MS/MS analysis of chloramphenicol
Samples were analyzed via reverse-phase liquid chromatography on an Agilent 1100 HPLC. The conditions were: Zorbax SB-C18 column, 3.0×150 mm (Agilent); mobile phase A: 25 mM ammonium acetate (pH = 6.8), mobile phase B: methanol; flow rate 0.5 mL/min, HPLC program as shown in Table S1. Compound 1 was quantified at 237 nm, while 4 was quantified at 285 nm by comparison with standards with known concentrations. An alternative HPLC method was used for in vivo assays, where buffer A (1% acetic acid) and buffer B (methanol) both contained 2 mM sodium 1-heptanesulfonic acid to improve peak shape and separation for the alternate substrates used. For mass spectroscopy work, an Agilent 1100 series LC/MSD XCT plus-ion-trap mass spectrometer was utilized. Identical mobile phases and columns were used as previously indicated, with a flow rate of 300 µL/min and the HPLC program shown below. The column effluent was directed to Agilent XCT ion-trap MSD mass spectrometer, which was operated in the negative ion mode to detect parent ions of each of the reaction intermediates. The system was operated using a drying temperature of 350 °C, a nebulizer pressure of 35 psi, a drying gas flow of 8.5 L/min, and a capillary voltage of 4500 V.
4.5. Ferrozine assay
The procedure was largely adapted from published protocol with slight modification.29 Briefly, 50 µL protein (20–200 µM range) was mixed with an equal volume of reducing reagent and incubated at room temperature for 5 min, followed by addition of 50 µL 12% trichloroacetic acid. After centrifugation, 100 µL supernatant was transferred to 40 µL 10% ammonium acetate buffer, followed by addition of 10 µL ferroin reagent. The mixture was incubated for 10 min at room temperature, and the absorbance was subsequently measured at 562 nm. Mohr’s salt was used to prepare the standard solutions, and molar extinction coefficient constant at 562 nm was 28,000 M−1 cm−1. The reducing reagent consisted of 48 µL A [ascorbic acid (4 mM), H2SO4 (0.1 M)] and 2 µL B [phenazine methosulfate (10 mM)]. The ferroin reagent was composed of ferrozine (6 mM) and H2SO4 (0.05 M).
Supplementary Material
Acknowledgements
This work was supported by a grant from the Office of Naval Research (N00014-02-1-0725) and Department of Chemical Engineering and Biomolecular Engineering at the University of Illinois at Urbana-Champaign (H.Z.). E.C. acknowledges support from the National Institutes of Health Molecular Biophysics Training Grant Program.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tet.2012.06.036.
References and notes
- 1.Winkler R, Hertweck C. ChemBioChem. 2007;8:973–977. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 2.Ju KS, Parales RE. Microbiol. Mol. Biol. Rev. 2010;74:250–272. doi: 10.1128/MMBR.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiwari MK, Lee JK, Moon HJ, Zhao H. Bioorg. Med. Chem. Lett. 2011;21:2873–2876. doi: 10.1016/j.bmcl.2011.03.087. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Zhao H. Angew. Chem. 2006;118:638–641. [Google Scholar]
- 5.Lee J, Simurdiak M, Zhao H. J. Biol. Chem. 2005;280:36719–36727. doi: 10.1074/jbc.M505334200. [DOI] [PubMed] [Google Scholar]
- 6.Simurdiak M, Lee J, Zhao H. ChemBioChem. 2006;7:1169–1172. doi: 10.1002/cbic.200600136. [DOI] [PubMed] [Google Scholar]
- 7.Choi YS, Zhang H, Brunzelle JS, Nair SK, Zhao H. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fries A, Bretschneider T, Winkler R, Hertweck C. ChemBioChem. 2011;12:1832–1835. doi: 10.1002/cbic.201100138. [DOI] [PubMed] [Google Scholar]
- 9.Winkler R, Richter MEA, Knüpfer U, Merten D, Hertweck C. Angew. Chem. Int. Ed. 2006;45:8016–8018. doi: 10.1002/anie.200603060. [DOI] [PubMed] [Google Scholar]
- 10.Zocher G, Winkler R, Hertweck C, Schulz GE. J. Mol. Biol. 2007;373:65–74. doi: 10.1016/j.jmb.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Vining LC, Studdard C. Genetics and Biochemistry of Antibiotics Production. Newton, Ma: Butterworth-Heinemann; 1995. [Google Scholar]
- 12.Westlake DWS, Vining LC. Biotechnol. Bioeng. 1969;11:1125–1134. doi: 10.1002/bit.260110609. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqueullah M, McGrath R, Vining LC. Can. J. Biochem. 1967;45:1881–1889. doi: 10.1139/o67-221. [DOI] [PubMed] [Google Scholar]
- 14.He J, Magarvey N, Piraee M, Vining LC. Microbiology. 2001;147:2817–2829. doi: 10.1099/00221287-147-10-2817. [DOI] [PubMed] [Google Scholar]
- 15.Piraee M, White RL, Vining LC. Microbiology (Reading, Engl.) 2004;150:85–94. doi: 10.1099/mic.0.26319-0. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Escribano JP, Bibb MJ. Micro Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacholec M, Sello JK, Walsh CT, Thomas MG. Org. Biomol. Chem. 2007;5:1692–1694. doi: 10.1039/b703356g. [DOI] [PubMed] [Google Scholar]
- 18.Makris TM, Chakrabarti M, Münck E, Lipscomb JD. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15391–15396. doi: 10.1073/pnas.1007953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanklin J, Achim C, Schmidt H, Fox BG, Münck E. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2981–2986. doi: 10.1073/pnas.94.7.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordlund P, Sjoberg BM, Eklund H. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K, Misawa M, Mitsuta Y. No. 03287546. Japan Patent. 1991
- 22.Corbett MD, Chipko BR. Antimicrob. Agents Chemother. 1978;13:193–198. doi: 10.1128/aac.13.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, Korboukh VK, Krebs C, Bollinger JM. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15722–15727. doi: 10.1073/pnas.1002785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korboukh VK, Li N, Barr EW, Bollinger JM, Krebs C. J. Am. Chem. Soc. 2009;131:13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramanian R, Rosenzweig AC. Acc. Chem. Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 27.Hakemian AS, Rosenzweig AC. Annu. Rev. Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 28.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 29.Paul C. Anal. Biochem. 1971;40:450–458. doi: 10.1016/0003-2697(71)90405-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.