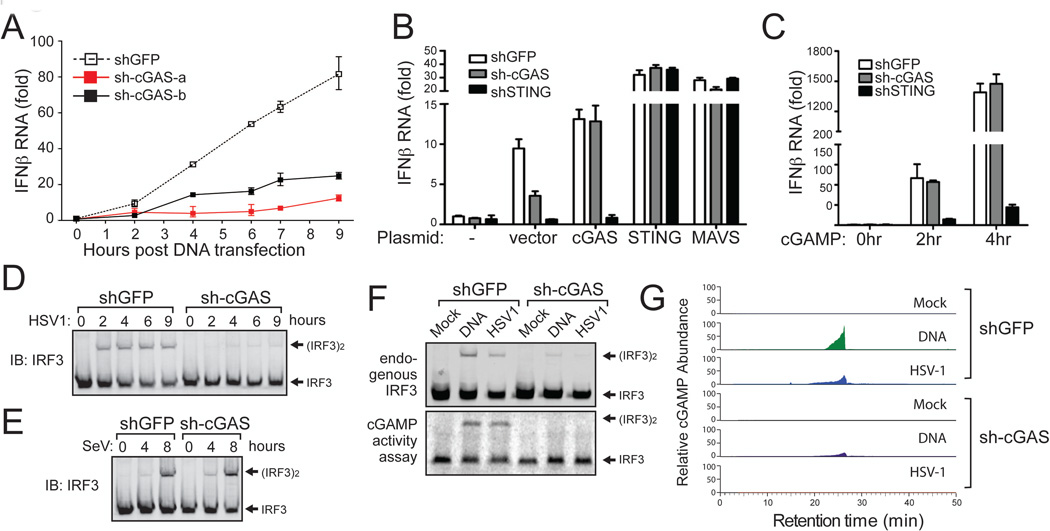
Figure 3. cGAS is essential for IRF3 activation and IFNβ induction by DNA transfection and DNA virus infection.
(A) L929 cell lines stably expressing shRNA targeting GFP (control) or two different regions of m-cGAS were transfected with HT-DNA for indicated times, followed by measurement of IFNβ RNA by q-RT-PCR. See fig. S4B for RNAi efficiency. (B) L929 cells stably expressing shRNA against GFP, cGAS or STING were transfected with pcDNA3 (vector) or the same vector driving the expression of indicated proteins. 24 hr after transfection, IFNβ RNA was measured by q-RT-PCR. see fig. S4C for RNAi efficiency. (C) cGAMP (100 nM) was delivered to digitoninpermeabilized L929/shRNA cells as indicated. IFNβ RNA was measured by q-RT-PCR at indicated times following cGAMP delivery. (D and E) L929-shRNA cells as indicated were infected with HSV1 (ΔICP34.5) or Sendai virus (SeV) for indicated times followed by measurement of IRF3 dimerization. (F) L929/shRNA cells were transfected with HT-DNA or infected with HSV1 for 6 hr, followed by measurement of IRF3 dimerization (top). Extracts from these cells were used to prepare heat-resistant supernatants, which were delivered to permeabilized Raw264.7 cells to stimulate IRF3 dimerization (bottom). (G) The heat-resistant supernatants in (F) were fractionated by HPLC using a C18 column and the abundance of cGAMP was quantitated by mass spectrometry using SRM.