Abstract
Women using menopausal hormone therapy (MHT) are at increased risk to develop breast cancer (BC). To detect genetic modifiers of the association between current use of MHT and BC risk, we conducted a meta-analysis of four genome-wide case-only studies followed by replication in eleven case-control studies. We used a case-only design to assess interactions between single nucleotide polymorphisms (SNPs) and current MHT use on risk of overall and lobular BC. The discovery stage included 2,920 cases (541 lobular) from four genome-wide association studies. The top 1,391 SNPs showing P-values for interaction (Pint) <3.0×10−03 were selected for replication using pooled case-control data from eleven studies of the Breast Cancer Association Consortium, including 7,689 cases (676 lobular) and 9,266 controls. Fixed effects meta-analysis was used to derive combined Pint. No SNP reached genome-wide significance in either the discovery or combined stage. We observed effect modification of current MHT use on overall BC risk by two SNPs on chr13 near POMP (combined Pint≤8.9×10−06), two SNPs in SLC25A21 (combined Pint≤4.8×10−05), and three SNPs in PLCG2 (combined Pint≤4.5×10−05). The association between lobular BC risk was potentially modified by one SNP in TMEFF2 (combined Pint≤2.7×10−05), one SNP in CD80 (combined Pint≤8.2×10−06), three SNPs on chr17 near TMEM132E (combined Pint≤2.2×10−06), and two SNPs on chr18 near SLC25A52 (combined Pint≤4.6×10−05). In conclusion, polymorphisms in genes related to solute transportation in mitochondria, transmembrane signaling and immune cell activation are potentially modifying BC risk associated with current use of MHT. These findings warrant replication in independent studies.
Keywords: breast cancer, genetic variation, menopausal hormone therapy, genome-wide
INTRODUCTION
Menopausal hormone therapy (MHT) is prescribed to women in order to alleviate climacteric symptoms and it is still commonly used despite evidence of associations with increased risk for cardiovascular diseases and breast cancer (Farquhar et al. 2009; Sprague et al. 2012; Tsai et al. 2011). Regarding breast cancer, only recent use of MHT increases risk and the elevated risk dissipates within two years after cessation of use (Narod 2011). Furthermore, the associated risk varies by type of MHT preparation and is greater for use of combined estrogen-progestogen therapy than for use of estrogen-monotherapy (Chlebowski and Anderson 2012; Narod 2011). A meta-analysis conducted in 2005 reported an odds ratio (OR) of 1.39 (95% confidence interval (CI) 1.12–1.72) for the association between use of combined estrogen-progestogen therapy and breast cancer risk, whereas the respective OR for use of estrogen-monotherapy was 1.16 (95% CI 1.06–1.28) (Shah et al. 2005). Also, differences by histology have been observed, with a stronger increase in risk for lobular and tubular breast cancer compared with ductal breast cancer (Bakken et al. 2011; Flesch-Janys et al. 2008).
Understanding of the role of female sex hormones in breast carcinogenesis has already led to the development of therapeutic strategies such as the adjuvant endocrine therapy for estrogen receptor positive breast cancer (Smith and Dowsett 2003). By investigating genetic modifiers of MHT associated breast cancer, the underlying mechanisms could be further elucidated. The detection of genes involved in hormone-related breast carcinogenesis could lead to new strategies for breast cancer prevention and treatment. Knowledge of genetic modifiers could also contribute to safer use of MHT as the individual risk to develop breast cancer when using MHT may vary depending on the genetic background.
Previous studies investigating interactions between single nucleotide polymorphisms (SNPs) and use MHT regarding breast cancer risk predominantly pursued a candidate gene approach. Most of the reported interactions have not been followed up in further studies (Hein et al. 2012; Justenhoven et al. 2012; Lee et al. 2011). The possible interaction with variants of the known genetic susceptibility loci for breast cancer in FGFR2 has not been clearly confirmed in subsequent studies (Campa et al. 2011; Kawase et al. 2009; Nickels et al. 2013; Prentice et al. 2009; Rebbeck et al. 2009; Travis et al. 2010). We previously failed to replicate the most significant interaction with MHT use observed for 2q36.3 in a genome-wide interaction study using case-only design (Hein et al. 2013).
We here expand our previous work (Hein et al. 2013) and conducted a meta-analysis of four case-only genome-wide gene-environment interaction studies for overall as well as for lobular breast cancer risk. We then evaluated the top 1,391 SNPs by case-control analyses utilizing data from eleven studies participating in the Breast Cancer Association Consortium (BCAC; http://ccge.medschl.cam.ac.uk/consortia/bcac/index.html).
MATERIALS AND METHODS
An overview of the included studies at each stage with respective numbers of cases and controls as well as the number of SNPs analyzed is displayed in Figure 1. All studies were approved by the relevant ethics committees and all participants gave informed consent.
Figure 1.
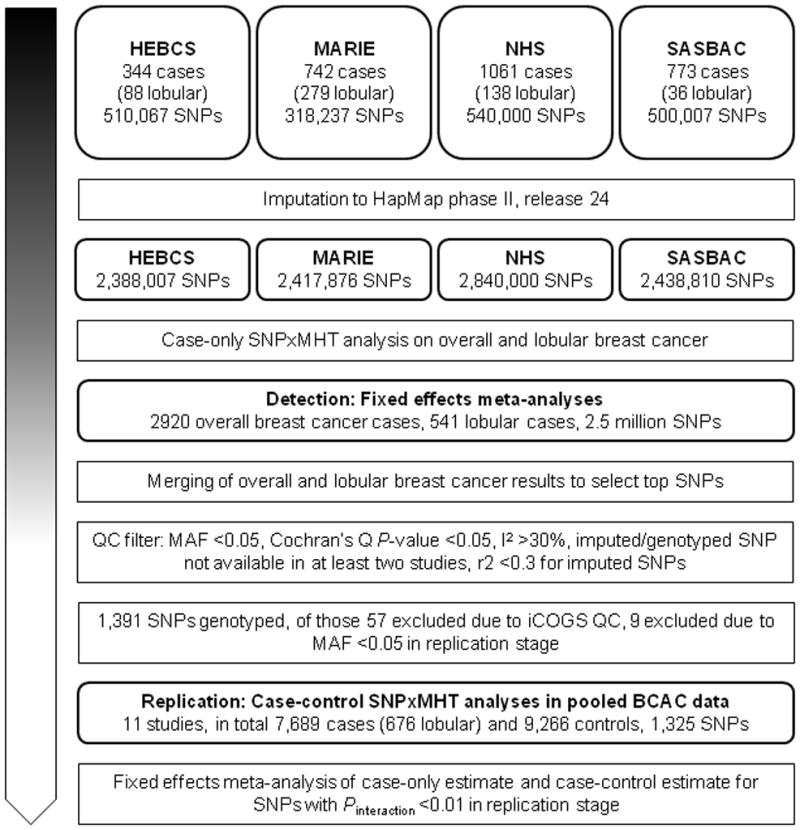
Diagram describing numbers of participants, investigated SNPs and conducted analyses at each stage
Study population of case-only genome-wide studies
Under the assumption that the genetic and environmental factors are not associated in the population from which the cases were drawn, case-only studies provide a powerful and efficient way to detect gene-environment interactions (Piegorsch et al. 1994). We conducted meta-analysis of four studies with quality control checked genome-wide data and information on current MHT use: the Mammary Carcinoma Risk Factor Investigation (MARIE) from Germany (Flesch-Janys et al. 2008), the Singapore and Sweden Breast Cancer Study (SASBAC) (Wedren et al. 2004), the Helsinki Breast Cancer Study (HEBCS) (Kilpivaara et al. 2004) and the Nurses’ Health Study (NHS) from the US (Hunter et al. 2007). Details on all studies participating in the discovery as well as replication stage can be found in Supplementary Table 1. In total these studies contributed 2,920 cases (541 cases with lobular tumors) to the meta-analysis.
Briefly, the MARIE study is a population-based case-control study of postmenopausal women aged 50–74 years carried out in two regions in Germany with incident cases diagnosed 2001–2005 and controls matched by birth year and study region (Flesch-Janys et al. 2008). Initially, 800 MARIE cases with known age at menopause were randomly selected for genotyping, with lobular cases oversampled (Hein et al. 2013). After quality control checks, a total of 742 MARIE cases were included in the case-only genome-wide association analysis, of which 279 were lobular cases. SASBAC is a subset of a Swedish nationwide population-based case-control study (Wedren et al. 2004). The cases were incident breast cancer cases diagnosed 1993–1995 identified via the six regional cancer registries in Sweden, to which reporting is mandatory. Overall, 773 cases (36 lobular tumors) were included in the case-only genome-wide analyses (Li et al. 2011). A further 344 postmenopausal cases (88 lobular) were contributed by the hospital-based Finnish study HEBCS. In HEBCS, cases included both unselected breast cancer and familial breast cancer patients recruited at the Helsinki University Central Hospital, 1997–2004 (Kilpivaara et al. 2004). The NHS cohort was established in 1976 and comprised 121,700 female registered nurses. In 1989–1990, 32,826 participants donated a blood sample. Of this sub-cohort 1,061 participants of European descent with incident postmenopausal invasive breast cancer (138 lobular) were included in the case-only genome-wide association analysis (Hunter et al. 2007). All subjects were of European ancestry.
Study populations used in the replication stage
SNPs selected from the case-only genome-wide association studies for replication were evaluated using seven population-based studies (five case-control studies (CECILE, GENICA, MARIE, PBCS, SASBAC), one case-cohort (MCCS), and one nested case-control study (UKBGS)), and four non-population-based studies (MCBCS, kConFab/AOCS, OFBCR, pKARMA), including in total 7,689 cases (676 lobular) and 9,266 controls participating in the Breast Cancer Association Consortium (BCAC). Studies from BCAC were included if participants were of European descent and if genotype information, information on MHT use and information on reference age was available for at least 200 postmenopausal cases and 200 postmenopausal controls. A reference age of ≥54 years was used as surrogate for defining postmenopausal status if study derived information on menopausal status was missing. Participants of SASBAC and MARIE were excluded if they contributed already to the respective case-only genome-wide association studies. Additionally, cases in MCCS and pKARMA with prevalent breast cancer at time of enrollment were excluded. The reference date for controls was date of enrollment (MCCS) or date of interview (case-control studies). The reference date for cases was the date of breast cancer diagnosis. The reference age was accordingly the age at reference date. In total, 7,698 cases (676 lobular) and 9,266 controls contributed to the replication analysis.
Menopausal hormone therapy exposure definition
Any type of MHT was taken into account when defining ever use of MHT. Only women using MHT more than three months were considered to be ever users. We defined current use of MHT as use within the last six months before reference date. Harmonization and plausibility checks of MHT information were conducted centrally for all studies participating in BCAC (Nickels et al. 2013).
Genotyping and Quality control
Genotyping was performed using the Illumina Humancnv370-duo chip (318,237 SNPs) in the MARIE study and the Illumina HumanHap550 chip I in SASBAC (500,007 SNPs), HEBCS (510,067 SNPs), and NHS (540,000 SNPs). Genotyping in NHS was part of the Cancer Genetic Markers of Susceptibility (CGEMS) project. All studies provided quality control checked genotype data.
SNPs selected for replication were genotyped on a custom Illumina iSelect genotyping array (iCOGS) that was designed by BCAC in collaboration with three other consortia (the Collaborative Oncological Gene-environment Study, COGS) (Michailidou et al. 2013). After genotyping the iCOGS data was centrally quality controlled, which lead to an exclusion of 56 SNPs selected for replication. We additionally excluded 9 SNPs with MAF < 0.05 in the replication dataset.
Imputation
All SNPs genotyped in the genome-wide studies, which were also contained in the HapMap phase II release 24 data, were used for imputation of additional genotypes using the software MACH 1.0.16 (Li et al. 2010). Employing the ‘autoflip’ option, the alleles are coded according to a unique reference scheme, so that the same allele was coded as reference in all four genome-wide case-only studies. For quality control of imputed data, imputed SNPs with minor allele frequency (MAF) <0.01 or r2 <0.3 were excluded from the analysis.
SNP selection for replication
A list of 1,391 SNPs was generated based on the lowest Pint (cut-off Pint <3.0×10−03) derived from the analysis of multiplicative interaction between MHT and breast cancer risk, after merging the results for overall and lobular breast cancer. A total of 3,277 SNPs were selected based on the interaction with overall breast cancer and 1,723 selected based on their association with lobular breast cancer. These SNPs were filtered according to the criteria MAF ≥0.05, P-value ≥0.05 for Cochran’s Q or I2 <30% and the availability of the respective SNP data in at least two case-only studies.
Statistical Analysis
We tested for multiplicative SNPxMHT interactions on the genome-wide level (2.5 million SNPs) in case-only analysis using logistic regression with MHT use (current use codes as 1, never/past use coded as 0) as the outcome variable and the SNP as the explanatory variable. The SNP was assessed according to a log-additive genetic model, i.e. a 1 df test for trend by number of minor alleles (0, 1, or 2). Uncertainty of imputed SNPs was accounted for by using estimated genotype probabilities for imputed SNPs in the regression model. Covariates were not considered in the case-only analyses. These analyses were performed with the software ProbABEL version 0.1–2-plus (Aulchenko et al. 2010). Only genotyped SNPs that were also contained in the HapMap reference data and imputed SNPs were included in case-only analyses. Analyses were performed for all cases as well as separately for lobular cases. Since only individuals of European descent were included, the genomic inflation factor lambda was close to one (HEBCS λ = 1.016; MARIE λ = 1.014; SASBAC λ = 1.009) and, in case of NHS, there was also no indication for population stratification (Hunter et al. 2007). Therefore, the analyses were not corrected for population stratification. Combined results based on the four case-only analyses were obtained from a meta-analysis assuming a fixed effects model, using the software PLINK, version 1.07 (Purcell et al. 2007). Heterogeneity between studies was assessed using Cochran’s Q statistic and I2 (Higgins and Thompson 2002).
For the replication analysis, data from eleven studies were pooled and analyzed using case-control logistic regression. These analyses were performed using SAS software, version 9.2. SNPxMHT interactions were evaluated by means of a log-likelihood ratio test, comparing models with and without a multiplicative interaction term between SNP (coded according to log-additive mode of inheritance) and current use of MHT. The models were adjusted for study, reference age, former use of MHT, and six principal components to account for population stratification. The models included also interaction terms between study design (non-population-based vs. population based) and current use of MHT as well as former use of MHT. These interaction terms were included to account for possible differences in the estimates of the MHT effect according to study design.
Results from the case-only meta-analysis and the replication were combined in a meta-analysis assuming a fixed effects model, using the package “meta”, version 2.1-2 (Schwarzer 2012) within the R software, version 2.15.0 (Team 2012). Linkage disequilibrium (LD) between selected SNPs was estimated in the control population of population-based studies using SNP_Tools, version 1.70 (Chen et al. 2009a) and Haploview, version 4.2 (Barrett et al. 2005).
The association between current MHT use and SNPs was assessed utilizing data of all studies of the replication stage as well as solely population-based studies. We fitted a logistic regression model adjusted for study with current MHT use as the outcome.
To illustrate the modification of overall as well as lobular breast cancer risk associated with current use of MHT by SNPs, the effect of current use of MHT was assessed in strata defined by the SNP genotype in pooled case-control data of the replication stage. The models were adjusted for study, reference age, former use of MHT and the two previously described interaction terms to account for possible differences in the estimates of the MHT effect according to study design.
To further evaluate the effect modification of current use of MHT by multiple modifying loci, a polygenic score was built for each individual. For the genetic score, we included the genetic loci found to modify the association with current use of MHT at a significance level of Pint <5.0×10−05 and selected one SNP per region based on the effect estimate. The allele increasing the effect of MHT on breast cancer risk was used as risk allele and the polygenic score was derived by summing up the risk alleles (0, 1 or 2) for each SNP. Separate scores were constructed for overall and lobular breast cancer risk. To demonstrate the polygenic modifying effect, associations of current MHT use with breast cancer risk were calculated stratified by score categories (roughly quintiles for all breast cancers and tertiles for lobular tumors). The respective logistic regression models were adjusted for study, reference age, former use of MHT, an interaction term between former use of MHT and study design (non-population-based vs. population-based) and an interaction term between current use of MHT and study design.
RESULTS
Characteristics of the patients participating in the studies included in the discovery stage and the replication stage are shown in Supplementary Table 2 and Supplementary Table 3. The prevalence of current use of MHT varied between 10% and 60%. The estimated OR for the marginal effect of current use of MHT in the population-based studies of the replication stage was 1.51 (95% CI 1.21 – 1.88) for overall breast cancer and 1.83 (95% CI 1.34 – 2.47) for lobular breast cancer, as displayed in Supplementary Figure 1.
The meta-analysis of the case-only genome-wide studies did not identify any SNPxMHT interaction at genome-wide significance level (see Supplementary Figure 2 and Supplementary Figure 3, respectively, for quantile-quantile (QQ) and Manhattan plots of the results). The strongest association was observed for rs3824418 located in and intronic region of TLE1 on 9q21.3 (Pint = 6.7×10−07.
Of the 1,391 SNPs carried forward for replication, 944 were selected based on their Pint for modification of overall breast cancer risk associated with MHT, and 447 based on their Pint for modification of lobular breast cancer risk. Overall, 7 SNPs showed effect modification of overall breast cancer with combined Pint <5.0×10−05, as well as 7 SNPs with respect to lobular breast cancer risk (Table 1). There was no strong association between the SNPs selected for follow-up and current use of MHT, as can be seen from QQ-plots in Supplementary Figure 4. When analyzing the whole sample of the replication stage, the SNPs showing strongest association with current use of MHT were rs12538442, located in an intronic region of DOCK4 on chromosome 1 (P = 3.1×10−04), and rs17738984, located on chromosome 17q22 (P = 8.8×10−04). The SNPs showing the strongest association with MHT use when restricting the sample to population-based studies were rs10924245 in KIF26B (P = 1.6×10−04) and rs2102354 in KCNN3 (P = 7.7×10−04), both located on chromosome 1.
Table 1.
Odds ratios of multiplicative interaction between current menopausal hormone therapy use and SNPs for overall and lobular breast cancer risk
| SNP | Chr | Position (build 37) | RefSeq Gene | Feature | GWASa | REPLICATION | COMBINEDc | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Pint | OR (95% CI)b | Pint | OR (95% CI) | Pint | |||||
| Overall breast cancer | ||||||||||
| rs9578047 | 13 | 29164731 | 68kb 5′ of POMP | - | 0.81 (0.72 – 0.91) | 6.3×10−04 | 0.84 (0.75 – 0.95) | 3.6×10−03 | 0.83 (0.76 – 0.90) | 7.9×10−06 |
| rs9579199 | 13 | 29164783 | 68kb 5′ of POMP | - | 0.81 (0.72 – 0.91) | 6.3×10−04 | 0.84 (0.75 – 0.95) | 3.4×10−03 | 0.83 (0.76 – 0.90) | 7.6×10−06 |
| rs7148646 | 14 | 37407036 | SLC25A21 | intronic | 0.80 (0.70 – 0.90) | 3.7×10−04 | 0.85 (0.76 – 0.96) | 7.4×10−03 | 0.83 (0.76 – 0.90) | 1.2×10−05 |
| rs848694 | 14 | 37371724 | SLC25A21 | intronic | 0.80 (0.70 – 0.91) | 9.4×10−04 | 0.86 (0.76 – 0.97) | 1.3×10−02 | 0.83 (0.76 – 0.91) | 4.6×10−05 |
| rs7192724 | 16 | 81958298 | PLCG2 | intronic | 1.24 (1.09 – 1.42) | 1.3×10−03 | 1.24 (1.09 – 1.40) | 7.8×10−04 | 1.24 (1.13 – 1.36) | 3.3×10−06 |
| rs17202296 | 16 | 81959191 | PLCG2 | intronic | 1.28 (1.13 – 1.46) | 1.4×10−04 | 1.14 (1.01 – 1.29) | 2.9×10−02 | 1.21 (1.10 – 1.32) | 2.8×10−05 |
| rs4888190 | 16 | 81963618 | PLCG2 | intronic | 1.32 (1.16 – 1.5) | 3.8×10−05 | 1.11 (0.99 – 1.26) | 8.0×10−02 | 1.20 (1.10 – 1.31) | 4.5×10−05 |
| Lobular breast cancer | ||||||||||
| rs11680872 | 2 | 192830249 | TMEFF2 | intronic | 2.01 (1.40 – 2.87) | 1.5×10−04 | 1.40 (1.05 – 1.87) | 1.9×10−02 | 1.61 (1.29 – 2.01) | 2.9×10−05 |
| rs7648642 | 3 | 119261375 | CD80 | intronic | 0.58 (0.43 – 0.79) | 4.6×10−04 | 0.68 (0.54 – 0.87) | 2.3×10−03 | 0.64 (0.53 – 0.78) | 5.1×10−06 |
| rs11654964 | 17 | 32989538 | 23kb 3′ of TMEM132E | 1.76 (1.30 – 2.38) | 2.7×10−04 | 1.58 (1.17 – 2.14) | 2.4×10−03 | 1.67 (1.35 – 2.06) | 2.7×10−06 | |
| rs16970162 | 17 | 32989786 | 23kb 3′ of TMEM132E | 1.85 (1.34 – 2.54) | 1.5×10−04 | 1.49 (1.09 – 2.05) | 1.1×10−02 | 1.66 (1.33 – 2.07) | 8.6×10−06 | |
| rs11080292 | 17 | 32998577 | 32kb 3′ of TMEM132E | 1.77 (1.29 – 2.44) | 4.6×10−04 | 1.57 (1.14 – 2.15) | 4.9×10−03 | 1.66 (1.33 – 2.08) | 9.3×10−06 | |
| rs6506940 | 18 | 29333635 | 6kb 3′ of SLC25A52 | 2.06 (1.35 – 3.13) | 7.2×10−04 | 1.80 (1.19 – 2.71) | 4.0×10−03 | 1.92 (1.43 – 2.58) | 1.2×10−05 | |
| rs594334 | 18 | 29364523 | 24kb 5′ of SLC25A52 | 1.92 (1.25 – 2.94) | 3.0×10−03 | 1.85 (1.22 – 2.81) | 3.0×10−03 | 1.88 (1.40 – 2.54) | 3.4×10−05 | |
fixed effects meta analysis of results from four case-only GWAS
adjusted for study, reference age, former use of menopausal hormone therapy, interaction terms between study design (population-bases vs. non-population-based) and former use and current of menopausal hormone therapy, as well as genetic principal components
fixed effects meta-analysis of results from GWAS and replication analysis
Further information on SNPs, including their association with overall breast cancer risk in the replication dataset and MAFs in the different study populations can be found in Supplementary Table 4. The identified SNPs on each of the chromosomes 13, 14, 16, 17 and 18 are located in close proximity to each other and do not represent independent signals of genetic modification of the MHT effect. The respective LD plots can be found in Supplementary Figure 5.
For overall breast cancer risk, two SNPs (rs9578047 and rs9579199) near POMP on chromosome 13 showed an interaction with current use of MHT with combined Pint = 7.9×10−06 and 7.6×10−06. SNPxMHT interactions with Pint of similar magnitude were also observed with rs7148646 and rs848694 in SLC25A21 on chromosome 14 (combined Pint = 1.2×10−05 and 4.6×10−05) and three SNPs (rs7192724, rs17202296, and rs4888190) in PLCG2 on chromosome 16 (combined Pint = 3.3×10−06, 2.8×10−05 and 4.5×10−05) (Table 1). Associations between current use of MHT and overall breast cancer risk stratified by genotypes of these SNPs are displayed in Table 2. We did not observe significantly heterogeneous SNPxMHT interactions between studies that were pooled in the replication stage (Phet ranging from 0.16 to 0.91). The respective forest plots are shown in Supplementary Figure 6.
Table 2.
Association between current use of menopausal hormone therapy and overall as well as lobular breast cancer risk stratified by genotype
| SNP | Allelesa | Homozygous reference | Heterozygous | Homozygous coded | ||||
|---|---|---|---|---|---|---|---|---|
| Reference | Coded | OR (95% CI)b | P | OR (95% CI)b | P | OR (95% CI)b | P | |
| Overall breast cancer | ||||||||
| rs9578047 | A | G | 1.91 (1.46 – 2.50) | 2.0×10−06 | 1.50 (1.30 – 1.72) | 1.3×10−08 | 1.30 (1.14 – 1.48) | 6.4×10−05 |
| rs9579199 | G | A | 1.92 (1.47 – 2.51) | 1.8×10−06 | 1.50 (1.30 – 1.72) | 1.4×10−08 | 1.30 (1.14 – 1.48) | 6.5×10−05 |
| rs7148646 | G | A | 1.58 (1.39 – 1.80) | 1.9×10−12 | 1.24 (1.08 – 1.43) | 2.6×10−03 | 1.31 (1.10 – 1.73) | 5.2×10−02 |
| rs848694 | G | A | 1.55 (1.37 – 1.75) | 3.2×10−12 | 1.23 (1.06 – 1.43) | 5.8×10−03 | 1.34 (0.97 – 1.85) | 7.5×10−02 |
| rs7192724 | C | G | 1.10 (0.78 – 1.54) | 5.9×10−01 | 1.25 (1.09 – 1.44) | 1.9×10−03 | 1.58 (1.40 – 1.80) | 7.8×10−13 |
| rs17202296 | G | T | 1.19 (0.88 – 1.62) | 2.5×10−01 | 1.32 (1.15 – 1.51) | 1.1×10−04 | 1.53 (1.35 – 1.74) | 7.0×10−11 |
| rs4888190 | C | G | 1.22 (0.89 – 1.66) | 2.1×10−01 | 1.36 (1.18 – 1.57) | 1.5×10−05 | 1.52 (1.34 – 1.73) | 1.3×10−10 |
| Lobular breast cancer | ||||||||
| rs11680872 | G | A | 1.33 (0.64 – 2.76) | 4.5×10−01 | 1.42 (1.04 – 1.94) | 2.6×10−02 | 2.19 (1.70 – 2.82) | 1.3×10−09 |
| rs7648642 | C | A | 3.05 (2.10 – 4.42) | 4.1×10−09 | 1.62 (1.23 – 2.14) | 6.0×10−04 | 1.43 (1.01 – 2.02) | 4.3×10−02 |
| rs11654964 | C | A | 0.24 (0.06 – 1.04) | 5.7×10−02 | 1.61 (1.18 – 2.19) | 2.8×10−03 | 2.15 (1.68 – 2.76) | 1.7×10−09 |
| rs16970162 | C | G | 0.33 (0.07 – 1.47) | 1.5×10−01 | 1.57 (1.14 – 2.17) | 6.3×10−03 | 2.09 (1.64 – 2.66) | 3.0×10−09 |
| rs11080292 | T | C | 0.38 (0.08 – 1.70) | 2.0×10−01 | 1.47 (1.06 – 2.03) | 2.1×10−02 | 2.15 (1.69 – 2.74) | 6.5×10−10 |
| rs6506940 | G | A | 0.94 (0.10 – 8.90) | 9.6×10−01 | 1.08 (0.72 – 1.61) | 7.2×10−01 | 2.05 (1.63 – 2.58) | 7.4×10−10 |
| rs594334 | C | T | 1.40 (0.24 – 7.98) | 7.1×10−01 | 1.03 (0.68 – 1.58) | 8.7×10−01 | 2.09 (1.67 – 2.61) | 1.6×10−10 |
the coded allele was not necessarily the minor allele.
adjusted for study, reference age, former use of menopausal hormone therapy, interaction between former use of menopausal hormone therapy and study design (non-population-based vs. population-based) and interaction between current use of menopausal hormone therapy and study design
We combined rs7148646_G, rs9579199_G, and rs7192724_G and constructed a polygenic score to assess breast cancer risk associated with current use of MHT depending on the modifying genetic risk (number of risk modifying alleles) (Figure 2A). For women with a low polygenic score of two or less risk modifying alleles (16.7% of women), current use of MHT was not associated with an increased breast cancer risk (OR = 1.04, 95% CI 0.87 – 1.25). Among women carrying three (30.9% of women) or four (34.2% of women) risk modifying alleles, current use of MHT was associated with a significantly increased breast cancer risk (OR = 1.25, 95% CI 1.08 – 1.45 and OR = 1.57, 95% CI 1.36 – 1.80, respectively). The strongest association between current use of MHT and breast cancer risk was observed for women with a polygenic score of five or six (18.2% of women) (OR = 1.86, 95% CI 1.56 – 2.22), as expected.
Figure 2.
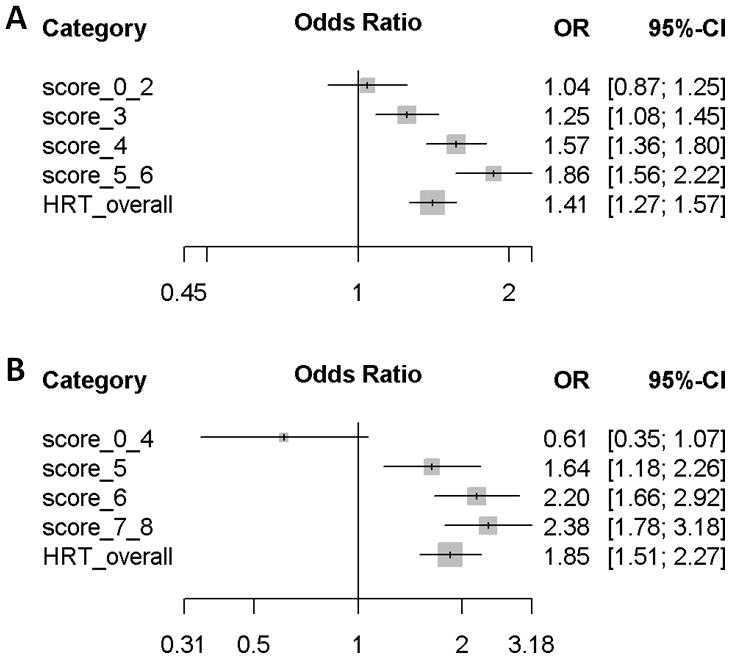
Odd ratios for (A) overall breast cancer and (B) lobular breast cancer risk associated with current use of menopausal hormone therapy in categories defined by polygenic scores
With respect to lobular breast cancer, the variant rs11680872 located in an intronic region of TMEFF2 on chromosome 2 showed a SNPxMHT interaction with combined Pint = 2.9×10−05 (Table 3). Also, rs7648642 in CD80 on chromosome 3 modified MHT associated lobular breast cancer risk in both case-only and case-control analysis (combined Pint = 5.1×10−06). The variants rs11654964, rs16970162 and rs11080292 located near TMEM132E on chromosome 17 yielded combined Pint of 2.7×10−06, 8.6×10−06 and 9.3×10−06, respectively. Further SNPxMHT interactions were observed for rs6506940 and rs594334 near SLC25A52 on chromosome 18 (combined Pint = 1.2×10−05 and 3.4×10−05, respectively) (Table 1). Table 2 shows the associations between current use of MHT and lobular breast cancer risk in strata defined by genotypes of these SNPs. There was no significant heterogeneity by study in the estimates for SNPxMHT interactions (Phet ranging from 0.22 to 0.94). The respective forest plots are shown in Supplementary Figure 7.
For lobular breast cancer risk, a polygenic score was constructed by combining rs11680872_A, rs7648642_C, rs11654964_A, and rs6506940_A (Figure 2B). Current use of MHT was not associated with increased lobular breast cancer risk in women carrying zero to four risk modifying alleles (15.2% of women, OR = 0.61, 95% CI 0.35 – 1.07), while the OR for lobular breast cancer risk was 1.64 (95% CI 1.18 – 2.26) in the subgroup (25.5% of women) carrying five risk modifying alleles. The association with current MHT use increased with the polygenic score and the odds ratio for lobular breast cancer was 2.20 (95% CI 1.66 – 2.92) in women with a polygenic score of six (30.9% of women) and 2.38, 95% CI 1.78 – 3.18) in those carrying seven or eight risk modifying alleles (25.8% of women).
DISCUSSION
Using a two stage approach consisting of a meta-analysis of four case-only genome-wide association studies and a replication analysis in independent data of ten case-control studies, we attempted to identify SNPs that modify the effect of MHT use on breast cancer risk. Despite our large sample size in both discovery and replication stages, we did not identify any SNPs that reached genome-wide significance. We observed three loci on chromosome 13, 14 and 16 that modified MHT associated overall breast cancer risk in both the discovery and replication stage, with combined Pint <5.0×10−05. Additionally, four genomic loci on chromosome 2, 3, 17 and 18 modified lobular breast cancer risk associated with MHT use in both study stages at the same significance level.
When combining variants that modify the effect of current use of MHT in a polygenic score, current MHT use was associated with an 86% increased breast cancer risk among women with five to six risk modifying alleles (18.2% of all women included in the study) and was not associated with breast cancer risk among women carrying two or less (16.7% of women). Women with a polygenic score of three or four that were currently using MHT were at an intermediate increased risk of breast cancer. Similar results were observed for lobular breast cancer risk. Given the observed interactions are confirmed, the polygenic score illustrates that even though the single detected interactions might be modest, the associated breast cancer risk for a woman using MHT may be appreciably increased if she carries a large number of adverse risk modifying alleles.
The loci of the identified polymorphisms provide indication of possible biological relevance for breast carcinogenesis. Two variants rs9579199 and rs9578047 close to FLT1 and POMP on chromosome 13 showed modifying effects on MHT associated breast cancer risk. FLT1 is a vascular endothelial growth factor receptor and involved in tumor angiogenesis (Fischer et al. 2008). So far, no association has been reported between tumor development and POMP, a proteasome maturation protein. The variants on chromosome 14 (rs7148646, rs848694) lie in an intronic region of SLC25A21. SLC25A21 encodes an oxodicarboxylate carrier, which transports C5–C7 oxodicarboxylates across the inner membranes of mitochondria (Fiermonte et al. 2001). Interestingly, the two SNPs rs6506940 and rs594334 found to modify risk of lobular breast cancer are located near another mitochondrial carrier gene, SLC25A52, on chromosome 18. Estrogen has been reported to be an important regulator of mitochondrial function (Chen et al. 2009b) and results of the present study suggest that mitochondrial related mechanisms may play a role in MHT associated breast carcinogenesis. The association between MHT and breast cancer was also modified by rs7192724, rs17202296 and rs4888190 located in intronic regions of PLCG2. PLCG2 is a member of the phosphoinositide-specific phospholipase C family and is involved in transmitting activation signals across the cell membranes predominantly of B cells (Wang et al. 2000) as well as natural killer cells (Tassi et al. 2005).
With respect to lobular breast cancer, genetic variants in two transmembrane proteins were implicated. rs11680872 is located in an intron of TMEFF2, whose biological function is unclear but its promoter region has been commonly found to be hypermethylated in various cancers, including breast cancer (Lin et al. 2011; Park et al. 2011). Three variants (rs11654964, rs16970162 and rs11080292) are near (23–32kb 3′) TMEM132E, another transmembrane protein. We observed also an interaction with rs7648642, which lies in an intron of CD80 on chromosome 3. CD80 is known to play an important role in T cell activation (Bhatia et al. 2006), and its expression has been found to be decreased in peripheral blood of breast cancer patients (Gong et al. 2012).
To account for differences by country with respect to types of preparations and dosages as well as different genotype platforms and laboratories, we used meta-analysis to combine the results in the discovery stage and adjusted for study in the replication stage. Heterogeneity of estimates regarding MHT use between studies due to differences in the study design were in part accounted for by adding an interaction term between MHT use and study design in the regression models (Supplementary Figure 1). The sensitivity analysis restricted to solely population-based studies of the replication stage for selected SNPxMHT interactions did not yield substantially altered estimates for interaction (less than 7.5% for overall breast cancer and less than 11.5% for lobular breast cancer, Supplementary Table 5). Utilizing the total study population of the replication stage, the study had 80% power to detect an interaction effect of 1.20, assuming an allele frequency of 20%, a marginal genetic effect of 1.15 and a marginal effect of current MHT use of 1.35. The power was reduced to 55% when restricting the sample to population-based studies. Furthermore, although the associations between current MHT use and breast cancer risk observed in the single studies were heterogeneous, this was not the case for the SNPxMHT interactions (Supplementary Figure 6 and 7). In general, estimates for gene-environment interaction are unlikely to be affected by selection bias (Morimoto et al. 2003) and more likely to be underestimated in the presence of non-differential or differential misclassification (Garcia-Closas et al. 1999).
Most of the reported genetic modifiers of MHT associated breast cancer risk have so far not been followed up in further studies (Justenhoven et al. 2012). One exception is with respect to variants in FGFR2, since it is also a know breast cancer susceptibility loci. Rebbeck et al. reported that the association between combined estrogen-progestogen therapy and breast cancer risk was modified by rs1219648 in postmenopausal women of European descent (Pint = 0.010) (Rebbeck et al. 2009). A study conducted in participants of the Women’s Health Initiative trial could not replicate this finding (Pint = 0.661) but observed an interaction with rs3750817 in FGFR2 (Pint = 0.033) (Prentice et al. 2009). A similar modifying effect of this SNP was also observed for estrogen-monotherapy (Pint = 0.046). The variants rs1219648 and rs3750817 are in moderate LD (r2 = 0.44, D′ = 1.00). However, we did not observe an interaction regarding breast cancer risk with current use of any MHT and rs1219648 (Pint = 0.15) or rs3750817 (Pint = 0.23) in the genome-wide association study and the variants were not followed up in the replication stage. Similarly, no significant interactions were observed with FGRR2 variants in more recent studies (Andersen et al. 2012; Campa et al. 2011; Nickels et al. 2013).
Genome-wide studies of gene-environment interactions present challenges since the required sample size maybe inflated due to misclassification of environmental exposures and additional factors involved including effect size of gene-environment interaction and prevalence of environmental exposure(s) (Dempfle et al. 2008; Zondervan and Cardon 2004). To optimize power, we used the case-only approach in the discovery stage, which offers greater precision in estimating the interaction term, and the case-control approach in the replication stage to account for false positive results due to correlation of the environmental exposure with the genetic marker in the population (Piegorsch et al. 1994). There was no indication of strong associations between SNPs selected for follow-up and current use of MHT (Supplementary Figure 4), supporting the assumption of gene-environment independence. We minimized possible spurious associations due to differences in allele frequencies in the underlying populations by restriction to solely individuals of European descent. The observed genomic inflation in the case-only studies was close to one and the case-control analyses were controlled for population stratification by including genetic principal components.
In conclusion, the association between current use of MHT and risk of overall and lobular breast cancer is potentially modified by genetic variants in genes related to mitochondrial solute carriers, transmembrane signaling as well as immune cell activation. These findings need replication in independent studies of adequate power. The identified modest interaction effects are presently unlikely to be of clinical significance, but provide valuable insight into potential mechanisms of breast cancer development.
Supplementary Material
Acknowledgments
FUNDING
Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME- ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund.
Meetings of the BCAC have been funded by the European Union COST programme (BM0606). D.F.E. is a Principal Research Fellow of CR-UK.
The CECILE study was funded by the Fondation de France; the French National Institute of Cancer (INCa); The National League against Cancer; the National Agency for Environmental and Occupational Health and Food Safety (ANSES), the National Agency for Research (ANR), and the Association for Research against Cancer (ARC).
The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114 as well as 01KH0401, 01KH0410, 01KH0411, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany.
The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I], the Hamburg Cancer Society, the German Cancer Research Center and genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402].
The MCBCS was supported by the NIH grants [CA122340, CA128978] and a Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201].
MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria.
The Nurses’ Health Studies are supported by US NIH (National Institute of Health) grants CA65725, CA87969, CA49449, CA67262, CA50385 and 5UO1CA098233.
OFBCR was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR.
The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA.
kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], the Cancer Council of Tasmania and Cancer Foundation of Western Australia and the NHMRC [199600]. ABS is supported by an NHMRC Senior Research Fellowship.
The Breakthrough Generations Study investigators thank Breakthrough Breast Cancer and the Institute of Cancer Research (ICR) for support and funding of the Breakthrough Generations Study. The ICR acknowledge NHS funding to the NIHR Biomedical Research Centre.
This study would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F. Easton, Paul Pharoah, Kyriaki Michailidou, Manjeet K. Bolla, Qin Wang (BCAC), Andrew Berchuck (OCAC), Rosalind A. Eeles, Douglas F. Easton, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Georgia Chenevix-Trench, Antonis Antoniou, Lesley McGuffog, Fergus Couch and Ken Offit (CIMBA), Joe Dennis, Alison M. Dunning, Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Javier Benitez, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. In particular, we thank: Ursula Eilber, Muhabbet Celik, Teresa Selander, Nayana Weerasooriya, Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner, The GENICA network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [Wing-Yee Lo, HB]; Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch]; Institute of Pathology, University of Bonn, Bonn, Germany [Hans-Peter Fischer]; Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [Ute Hamann]; Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany [TB, Beate Pesch, Sylvia Rabstein, Anne Lotz] and Institute for Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [VH], Heather Thorne, Eveline Niedermayr and the kConFab Clinical Follow-Up Study, the AOCS Management Group (D. Bowtell, G. Chenevix-Trench, A. deFazio, D. Gertig, A. Green, P. Webb), the ACS Management Group (A. Green, P. Parsons, N. Hayward, P. Webb, D. Whiteman); The Australian Cancer Study Management Group (A. Green, P. Parsons, N. Hayward, P.M.Webb, and D. Whiteman), project staff, collaborating institutions, clinical and scientific collaborators and study participants.
Footnotes
DECLARATION OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Andersen SW, Trentham-Dietz A, Figueroa JD, Titus LJ, Cai Q, Long J, Hampton JM, Egan KM, Newcomb PA. Breast cancer susceptibility associated with rs1219648 (fibroblast growth factor receptor 2) and postmenopausal hormone therapy use in a population-based United States study. Menopause. 2012 doi: 10.1097/GME.0b013e318268ca46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken K, Fournier A, Lund E, Waaseth M, Dumeaux V, Clavel-Chapelon F, Fabre A, Hemon B, Rinaldi S, Chajes V, et al. Menopausal hormone therapy and breast cancer risk: impact of different treatments. The European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2011;128:144–156. doi: 10.1002/ijc.25314. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Edidin M, Almo SC, Nathenson SG. B7-1 and B7-2: similar costimulatory ligands with different biochemical, oligomeric and signaling properties. Immunol Lett. 2006;104:70–75. doi: 10.1016/j.imlet.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wilkening S, Drechsel M, Hemminki K. SNP_tools: A compact tool package for analysis and conversion of genotype data for MS-Excel. BMC Res Notes. 2009a;2:214. doi: 10.1186/1756-0500-2-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim Biophys Acta. 2009b;1793:1540–1570. doi: 10.1016/j.bbamcr.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Anderson GL. Changing concepts: Menopausal hormone therapy and breast cancer. J Natl Cancer Inst. 2012;104:517–527. doi: 10.1093/jnci/djs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempfle A, Scherag A, Hein R, Beckmann L, Chang-Claude J, Schafer H. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16:1164–1172. doi: 10.1038/ejhg.2008.106. [DOI] [PubMed] [Google Scholar]
- Farquhar C, Marjoribanks J, Lethaby A, Suckling JA, Lamberts Q. Long term hormone therapy for perimenopausal and postmenopausal women. Cochrane Database Syst Rev. 2009:CD004143. doi: 10.1002/14651858.CD004143.pub2. [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, Palmieri L, Ventura M, Runswick MJ, Palmieri F, Walker JE. Identification of the human mitochondrial oxodicarboxylate carrier. Bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J Biol Chem. 2001;276:8225–8230. doi: 10.1074/jbc.M009607200. [DOI] [PubMed] [Google Scholar]
- Fischer C, Mazzone M, Jonckx B, Carmeliet P. FLT1 and its ligands VEGFB and PlGF: drug targets for anti-angiogenic therapy? Nat Rev Cancer. 2008;8:942–956. doi: 10.1038/nrc2524. [DOI] [PubMed] [Google Scholar]
- Flesch-Janys D, Slanger T, Mutschelknauss E, Kropp S, Obi N, Vettorazzi E, Braendle W, Bastert G, Hentschel S, Berger J, et al. Risk of different histological types of postmenopausal breast cancer by type and regimen of menopausal hormone therapy. Int J Cancer. 2008;123:933–941. doi: 10.1002/ijc.23655. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Rothman N, Lubin J. Misclassification in case-control studies of gene-environment interactions: assessment of bias and sample size. Cancer Epidemiol Biomarkers Prev. 1999;8:1043–1050. [PubMed] [Google Scholar]
- Gong J, Pan W, Yang C, Guo F, Sun Y. Expression of co-stimulators CD28/B7-1 in peripheral blood of patients with breast cancer. Breast Cancer Res Treat. 2012;136:621–622. doi: 10.1007/s10549-012-2267-2. [DOI] [PubMed] [Google Scholar]
- Hein R, Abbas S, Seibold P, Salazar R, Flesch-Janys D, Chang-Claude J. Polymorphism Thr160Thr in SRD5A1, involved in the progesterone metabolism, modifies postmenopausal breast cancer risk associated with menopausal hormone therapy. Breast Cancer Res Treat. 2012;131:653–661. doi: 10.1007/s10549-011-1772-z. [DOI] [PubMed] [Google Scholar]
- Hein R, Flesch-Janys D, Dahmen N, Beckmann L, Lindstrom S, Schoof N, Czene K, Mittelstrass K, Illig T, Seibold P, et al. A genome-wide association study to identify genetic susceptibility loci that modify ductal and lobular postmenopausal breast cancer risk associated with menopausal hormone therapy use: a two-stage design with replication. Breast Cancer Res Treat. 2013 doi: 10.1007/s10549-013-2443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justenhoven C, Obazee O, Brauch H. The pharmacogenomics of sex hormone metabolism: breast cancer risk in menopausal hormone therapy. Pharmacogenomics. 2012;13:659–675. doi: 10.2217/pgs.11.144. [DOI] [PubMed] [Google Scholar]
- Kawase T, Matsuo K, Suzuki T, Hiraki A, Watanabe M, Iwata H, Tanaka H, Tajima K. FGFR2 intronic polymorphisms interact with reproductive risk factors of breast cancer: results of a case control study in Japan. Int J Cancer. 2009;125:1946–1952. doi: 10.1002/ijc.24505. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111:543–547. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- Lee E, Schumacher F, Lewinger JP, Neuhausen SL, Anton-Culver H, Horn-Ross PL, Henderson KD, Ziogas A, Van Den Berg D, Bernstein L, et al. The association of polymorphisms in hormone metabolism pathway genes, menopausal hormone therapy, and breast cancer risk: a nested case-control study in the California Teachers Study cohort. Breast Cancer Res. 2011;13:R37. doi: 10.1186/bcr2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Humphreys K, Heikkinen T, Aittomaki K, Blomqvist C, Pharoah PD, Dunning AM, Ahmed S, Hooning MJ, Martens JW, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using Sequence and Genotype Data to Estimate Haplotypes and Unobserved Genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Taylor JR, Jr, Wu TD, Gutierrez J, Elliott JM, Vernes JM, Koeppen H, Phillips HS, de Sauvage FJ, Meng YG. TMEFF2 is a PDGF-AA binding protein with methylation-associated gene silencing in multiple cancer types including glioma. PLoS One. 2011;6:e18608. doi: 10.1371/journal.pone.0018608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013 doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto LM, White E, Newcomb PA. Selection bias in the assessment of gene-environment interaction in case-control studies. Am J Epidemiol. 2003;158:259–263. doi: 10.1093/aje/kwg147. [DOI] [PubMed] [Google Scholar]
- Narod SA. Hormone replacement therapy and the risk of breast cancer. Nat Rev Clin Oncol. 2011;8:669–676. doi: 10.1038/nrclinonc.2011.110. [DOI] [PubMed] [Google Scholar]
- Nickels S, Truong T, Hein R, Stevens K, Buck K, Behrens S, Eilber U, Schmidt M, Haberle L, Vrieling A, et al. Evidence of Gene-Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors. PLoS Genet. 2013;9:e1003284. doi: 10.1371/journal.pgen.1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, Kim IA, Jung N, Cho NY, Kang GH. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Huang Y, Hinds DA, Peters U, Pettinger M, Cox DR, Beilharz E, Chlebowski RT, Rossouw JE, Caan B, et al. Variation in the FGFR2 gene and the effects of postmenopausal hormone therapy on invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3079–3085. doi: 10.1158/1055-9965.EPI-09-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, Strom BL. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30:269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer G. meta: Meta-Analysis with R 2012 [Google Scholar]
- Shah NR, Borenstein J, Dubois RW. Postmenopausal hormone therapy and breast cancer: a systematic review and meta-analysis. Menopause. 2005;12:668–678. doi: 10.1097/01.gme.0000184221.63459.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348:2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- Sprague BL, Trentham-Dietz A, Cronin KA. A Sustained Decline in Postmenopausal Hormone Use Results From the National Health and Nutrition Examination Survey, 1999–2010. Obstet Gynecol. 2012;120:595–603. doi: 10.1097/AOG.0b013e318265df42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi I, Presti R, Kim S, Yokoyama WM, Gilfillan S, Colonna M. Phospholipase C-gamma 2 is a critical signaling mediator for murine NK cell activating receptors. J Immunol. 2005;175:749–754. doi: 10.4049/jimmunol.175.2.749. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, Beral V, Peto R, Bell J, Zelenika D, et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375:2143–2151. doi: 10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SA, Stefanick ML, Stafford RS. Trends in menopausal hormone therapy use of US office-based physicians, 2000–2009. Menopause. 2011;18:385–392. doi: 10.1097/gme.0b013e3181f43404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Feng J, Wen R, Marine JC, Sangster MY, Parganas E, Hoffmeyer A, Jackson CW, Cleveland JL, Murray PJ, et al. Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity. 2000;13:25–35. doi: 10.1016/s1074-7613(00)00005-4. [DOI] [PubMed] [Google Scholar]
- Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, et al. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–449. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


