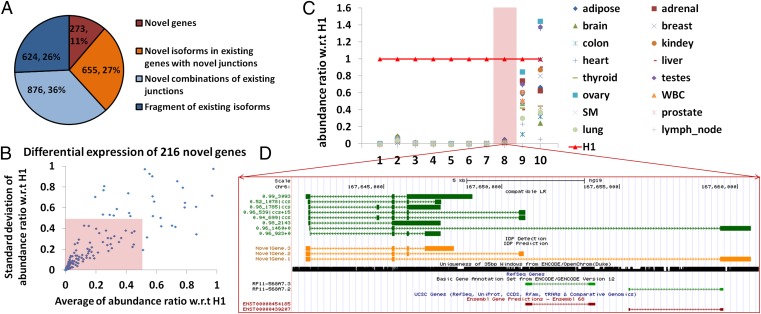
Fig. 2.
Novel gene identifications. (A) A total of 2,428 novel isoforms are categorized according to the use of the annotated junctions. A total of 273 isoforms from 216 novel genes are observed (in brown). A total of 655 novel isoforms use at least one junction from annotated genes (in orange). A total of 876 novel isoforms are novel combinations of annotated junctions. Six hundred twenty-four are fragments of annotated isoforms. (B) Differential expression of 216 novel genes in H1. The abundance ratio of a novel gene in a given tissue is defined as its abundance in this tissue divided by its abundance in H1. One hundred forty-six novel genes (68%, inside the pink box) have an averaged abundance (among 16 human tissues) ratio smaller than 0.5 with SD smaller than 0.5. (C) Relative expression levels of the top 10 novel genes (10 highest expressions in hESCs) in 16 human tissues. The reference expression levels are expressions in hESCs (highlighted in red line with triangles). Eight novel genes have high expression specifically in hESCs (example 8 in D) whereas the other 2 have significant expression across many different tissues. The gene structure of the eighth novel gene is visualized in D. (D) Novel gene at chr6:167,641,267–167,660,912. The dark green track shows the nonredundant long reads, each of which represents an alignment. The arrow refers to the alignment of the read relative to the reference (i.e., aligned to reference or to reverse complement of reference) and is not the direction of transcription. The naming of nonredundant long reads is A_B|ccs ± D, where A is the percentage identity of BLAT alignment, B is the length of alignment, and D is the distance between the mappable part of the long read and the polyA/polyT detection (“+” is the forward strand and downstream whereas “−” is the reverse strand and upstream). PacBio circular consensus sequence (CCS) reads are labeled with “ccs”. The orange track shows IDP predictions. The 35-bp mappability of this locus is in black. The light green track is GENCODE annotation and the brown one is Ensembl. RefSeq (light purple track) and UCSC Genes are also displayed but they have no annotated genes in this locus and thus no IDP detections (referenced to RefSeq, red track) are displayed either. The track display settings of other figures are the same.