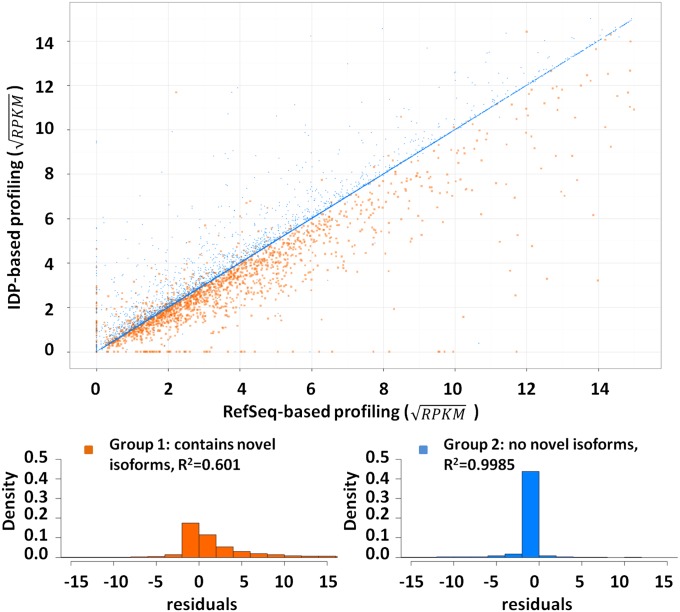
Fig. 7.
Isoform abundance estimation by IDP-identified hESC transcriptome and RefSeq annotation. The gene abundances of “common isoforms” (main text) are rescaled by a square-root transformation. The genes without novel isoforms (group 2) have stable abundance estimation and have high R2 of 0.9985 in linear regression (blue dots). In contrast, novel isoforms found in existing genes lead to a large range of abundance corrections. The residuals of linear regression show different distributions between the two groups. Group 1 (genes with novel isoforms) is highlighted in orange and group 2 in blue. The residuals of group 2 concentrate around 0, which indicates a small difference between two computations. However, group 1 has a heavy tail at the positive range. That is, most abundances of group 1 are corrected to lower values because the SGS reads must be shared with novel isoforms.