FIG. 1.
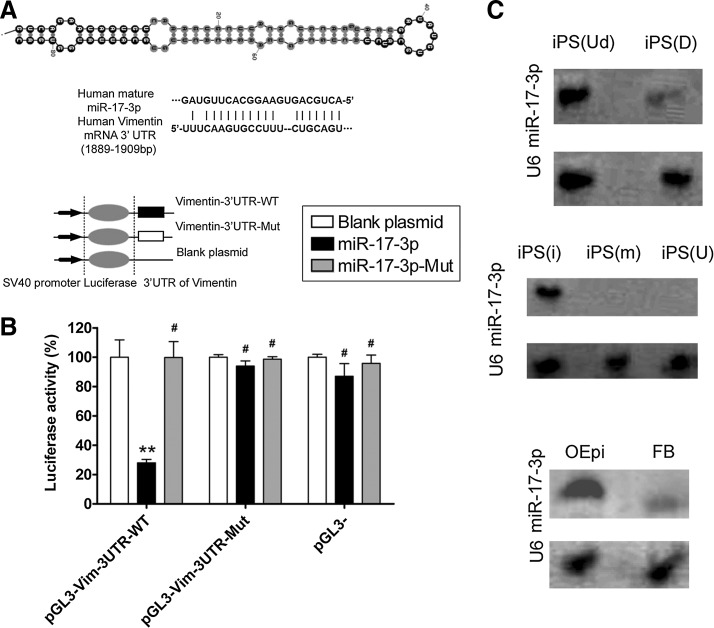
Secondary structure of miR-17-3p and its expression in different cells (A) Typical secondary structure of hsa-miR-17-3p precursor miRNAs (pre-miRNAs), contains stem-loop and hairpin structures, with a binding site located in an unstable region with a multi-branching loop-like RNA structure. Complementarity between miR-17-3p and the targeted human vimentin 3′-UTR site (1889–1909 bp downstream), and conserved bases of the mature miR-17-3p target sequence were present in the human vimentin 3′-UTR. (B) Expression of miR-17-3p and its effect on expression of the target gene vimentin was assessed by a luciferase assay. Wild-type (WT) reporter or mutated control luciferase plasmids were transfected into NIH-3T3 cells with miR-17-3p or mut-miR-17-3p expressing lentiviruses. Luciferase activity within the vimentin 3′-UTR sites was inhibited by miR-17-3p (**p<0.01 vs. empty plasmid; #p>0.05 vs. empty plasmid; n=3). (C) Northern blotting results showing expression of miR-17-3p in different cells. In induced pluripotent stem (iPS) cells, a stronger miR-17-3p hybridization signal was observed in human iPS cells at the differentiated stage (D), compared with that at the undifferentiated stage (Ud). In transfected iPS cells, a stronger miR-17-3p hybridization signal was observed in human iPS cells transfected with the miR-17-3p expression plasmid (i), compared with those that were transfected with the mutant miR-17-3p (m) expression plasmid or were untransfected (U). In differentiated iPS cells, a stronger miR-17-3p hybridization signal was seen in ovarian epithelial-like cells derived from iPS cells (OEpi), compared with that in fibroblast cells (FB). U6 RNA was used as a loading control in northern blot assays.