Figure 1.
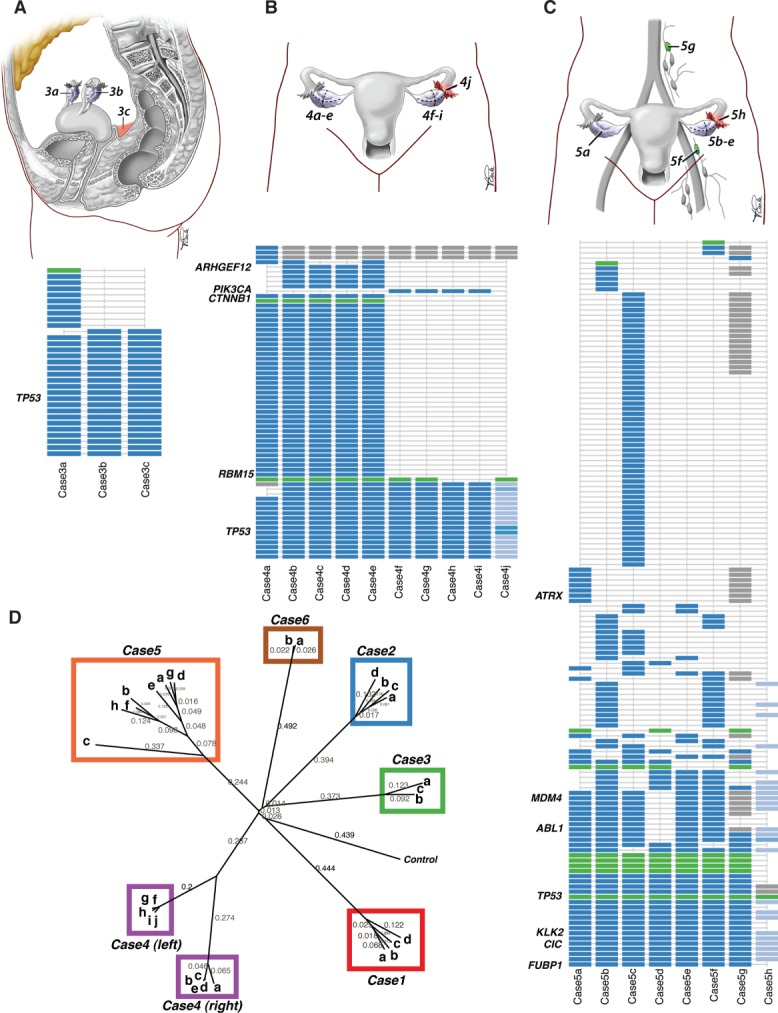
Intratumoural mutational profiles of HGS ovarian cancer. Anatomical sites and intratumoural mutational profile for cases 3 (A), 4 (B) and 5 (C); point mutations are shown in blue, indels in green; grey indicates a predicted mutation where validation by deep sequencing was inconclusive; light blue indicates allelic frequencies (counts of non-reference allele/total depth of coverage) in the fallopian tube lesion. (D) Phylogenetic tree of mutational profiles of cases 1–6, depicting evolutionary branching patterns reflective of clonal relationships between samples. The tree was computed using distance matrices based on Pearson correlation coefficients, followed by neighbour-joining cluster analysis. The control sample represents the ‘root’ whereby data were generated with no aberrations. Neighbour-joining distances are shown along the branches of the tree, which reflect genetic distances between branching points; longer branches indicate more genomic differences.
