Abstract
When compared to other conserved housekeeping protein families, such as ribosomal proteins, during the evolution of higher eukaryotes, aminoacyl-tRNA synthetases (aaRSs) show an apparent high propensity to add new sequences, and especially, new domains. The stepwise emergence of those new domains is consistent with their involvement in a broad range of biological functions beyond protein synthesis, and correlates with the increasing biological complexity of higher organisms. The new domains have been extensively characterized based on their evolutionary origins and their sequence, structural and functional features. While some of the domains are uniquely found in aaRSs and may have originated from nucleic acid binding motifs, others are common domain modules mediating protein-protein interactions that play a critical role in the assembly of the multi-synthetase complex (MSC). Interestingly, the MSC has emerged from a miniature complex in yeast, to a large, stable complex in insects to humans. The human MSC consists of 9 aaRSs (LysRS, ArgRS, GlnRS, AspRS, MetRS, IleRS, LeuRS and GluProRS) and 3 scaffold proteins (AIMP1/p43, AIMP2/p38 and AIMP3/p18), and has a molecular weight of 1.5 million Da. The MSC has been proposed to have a functional dualism: both facilitating protein synthesis and serving as a reservoir of non-canonical functions associated with its synthetase and non-synthetase components. Importantly, domain additions and functional expansions are not limited to the components of the MSC and are found in almost all aaRS proteins. From a structural perspective, multi-functionalities are represented by multiple conformational states. In fact, alternative conformations of aaRSs have been generated by various mechanisms from proteolysis to alternative splicing and posttranslational modifications, as well as by disease-causing mutations. Therefore, the metamorphosis between different conformational states is connected to the activation and regulation of the novel functions of aaRSs in higher eukaryotes.
Keywords: Domain expansion, Evolution, Novel function, Structural metamorphosis, tRNA synthetase
1 Introduction
The first surprise of the human genome project was the discovery of an unexpectedly small number of protein-coding genes [1]. The explanation that has slowly emerged since is that human genes possess more diverse functions and their regulation is more complicated than their counterpart genes in lower organisms. In this regard, the increasing complexity of aminoacyl-tRNA synthetases (aaRSs)—at the sequence, structural and functional levels—stands out as a prominent example.
Known as an essential component of the translational apparatus, the aaRS family catalyzes the first step reaction in protein synthesis, that is to specifically attach each amino acid to its cognate tRNA. While preserving this essential role, higher eukaryotic tRNA synthetases have developed other roles during evolution. Human cytoplasmic tRNA synthetases, in particular, mediate diverse functions in different pathways including angiogenesis, inflammation, development and tumorigenesis. The functional expansion of aaRSs is thought to be intimately associated with their continuous addition of new domains and motifs during the evolution of higher eukaryotes. Here we review the current knowledge on how human cytoplasmic tRNA synthetases developed their complexity, in sequence and in structure, to expand their “functionome”. Importantly, not all conserved housekeeping protein families possess the increasing complexity of the aaRS family.
2 Stepwise Appearance of New Domains and Sequences in Higher Eukaryotes
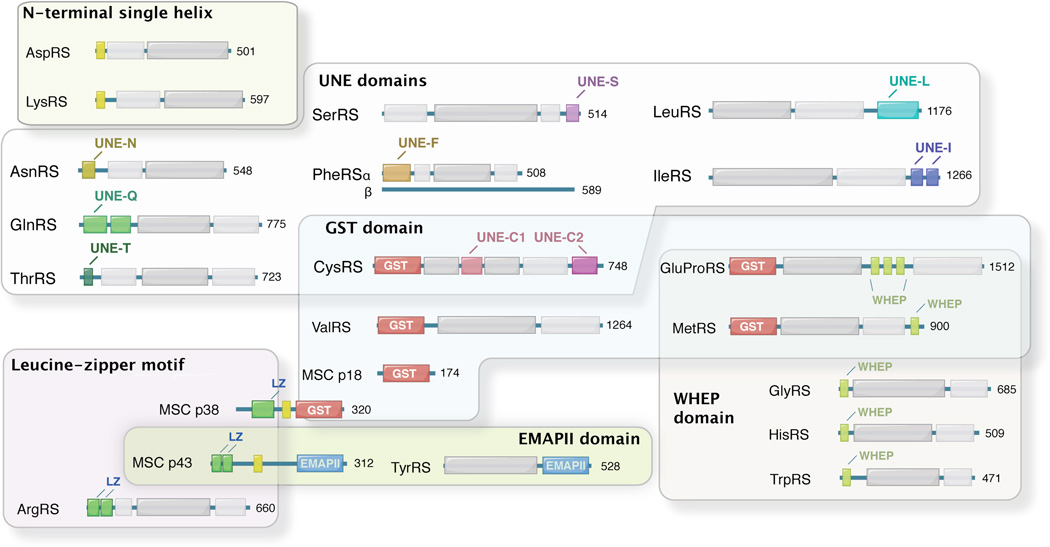
Comparing the protein sequences of eukaryotic cytoplasmic aaRSs with their prokaryotic counterparts, it is immediately obvious that eukaryotic cytoplasmic aaRSs are generally larger [2], which is mainly due to extensions or insertions of conserved sequences (Figure 1). Interestingly, most of these additions are found, at least by structural predictions, to form well-folded structures. Some of them are homologous to structural modules such as leucine-zipper and glutathione S-transferase (GST) domains that are broadly contained in human proteins, whereas others are only found in aaRSs or aaRS-associated protein factors. Those aaRS-specific domains include two common domains (WHEP and EMAPII) that are found in more than one member of the aaRS protein family, and several unique domains that each is found in a single aaRS. In addition to those well-folded domains, some shorter sequences that may not form defined structures are also found as appendices in eukaryotic aaRSs (Figure 1).
Figure 1.
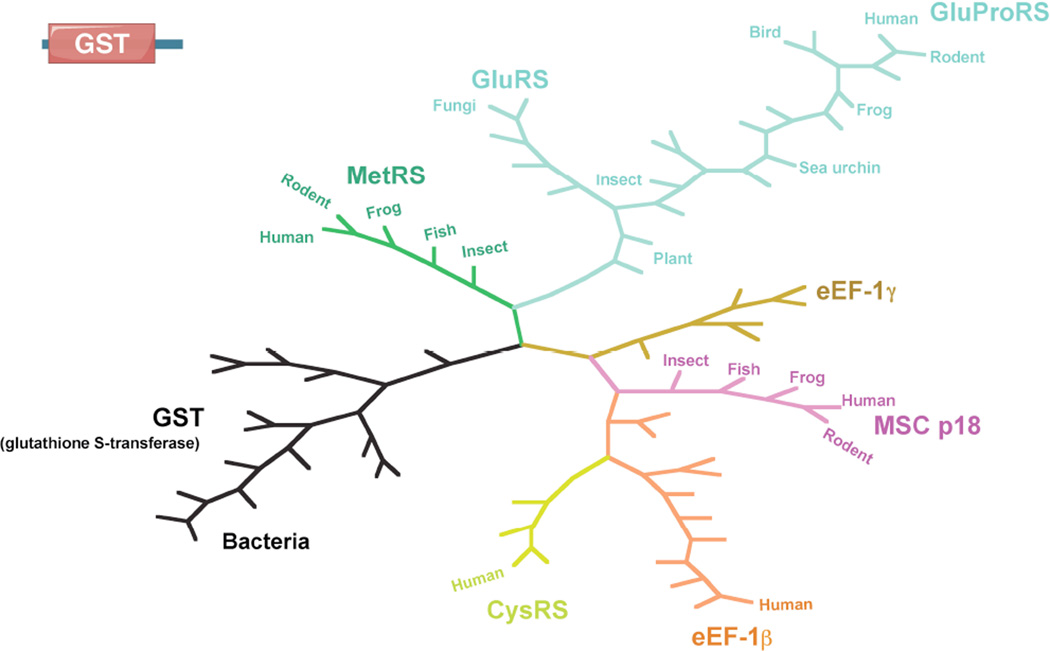
New domains and sequence additions as found in human aaRSs.
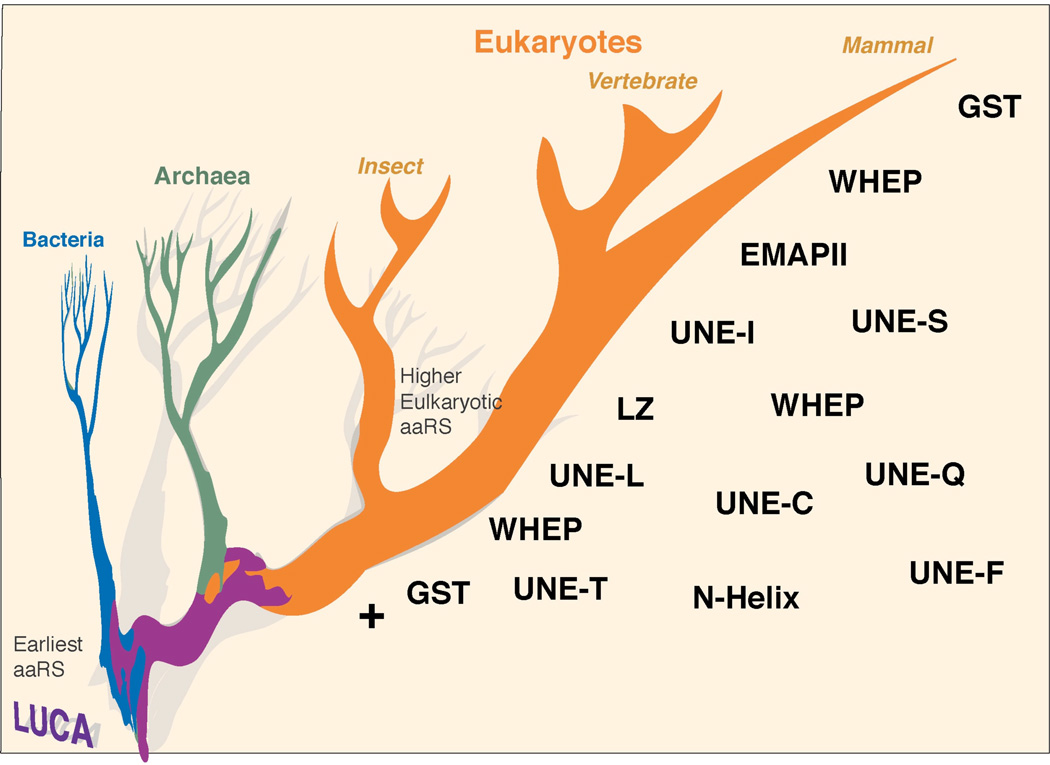
Remarkably, the elaboration of such new domains in aaRS proteins reflects the increasing biological complexity of the higher organisms wherein the proteins reside. The more evolutionarily advanced an organism, the more of those domains and sequences appears in its aaRS proteins. Therefore, the evolutionary process to add those domains and sequences on aaRSs appears to be continuous and almost irreversible (Figure 2).
Figure 2.
Elaboration of new domains and sequence additions on aaRSs during the evolution of eukaryotes.
2.1 N-terminal Helix
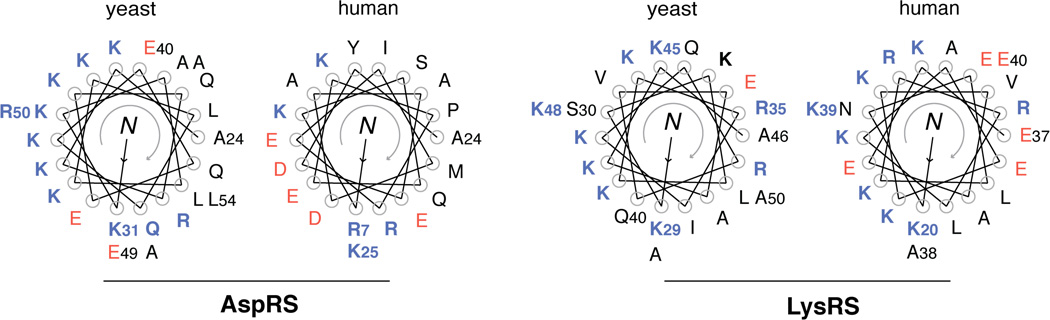
The simplest form of a eukaryotic domain addition is the N-terminal helix. Single helix sequences are found in the N-terminal region of eukaryotic LysRS and AspRS. These two aaRSs belong to the Class IIb of aaRS family and are more closely related to each other than to other aaRS members. Prokaryotic AspRS and LysRS consist only an OB (oligonucleotide binding)-fold tRNA anticodon-binding domain and an aminoacylation domain. However, during evolution of single-celled eukaryote yeasts to humans, each of the two synthetases acquired a ∼30–50 aa long N-terminal extension in front of the tRNA binding domain. The extensions, as suggested by structural prediction, contain a long helix of 20–40 aa (Figure 3). The helical conformation was confirmed by NMR structure determination of the N-terminal extensions of human AspRS and human LysRS [3, 4].
Figure 3.
Evolution of N-helix in eukaryotic AspRS and LysRS.
These N-terminal helices have evolved to mediate biological activities. They are mostly amphiphilic, with charged residues on one side and hydrophobic residues on the other side. For LysRS, positively charged residues dominate the hydrophilic side of the N-terminal helix. Early work indicated that this helical extension in human LysRS binds to the elbow region of tRNALys and enhances the binding affinity of the synthetase for the tRNA [5, 6]. Human LysRS also plays an important role in HIV infection by delivering tRNALys3 (which acts as a primer for viral reverse transcription) into the virion. Importantly, the function of LysRS in HIV packaging depends on the N-terminal extension, presumably because of its tRNA binding property [7].
Interestingly, the positively charged N-terminal helix of human LysRS not only interacts with nucleic acids but also with phospholipids and proteins. Recent work has shown that human LysRS, upon phosphorylation, is translocated to the plasma membrane where its N-terminal region, including the N-terminal extension, interacts with the transmembrane region of 67LR laminin receptor. The interaction inhibits the ubiquitin-dependent degradation of 67LR thereby enhancing laminin-induced cancer cell migration [8].
For yeast AspRS, the N-terminal helix is also positively charged on one side, and has been demonstrated to have tRNA binding property [9]. Interestingly, compared to the yeast enzyme, human AspRS possesses more negatively charged residues located on the N-terminal helix (Figure 3), suggesting that the amphiphilic helices may have evolved from facilitating tRNA binding in lower eukaryotes to mediating other interactions in higher eukaryotes [10–12].
2.2 EMAPII Domain
Another appended domain that facilitates tRNA binding is the EMAPII domain. Although originally discovered as a cytokine (endothelial monocyte activating polypeptide II), it is in fact a proteolytic product of MSC p43/AIMP1 – a scaffold protein in the multi-aminoacyl-tRNA synthetase complex (MSC; see below for more information on MSC) [13].
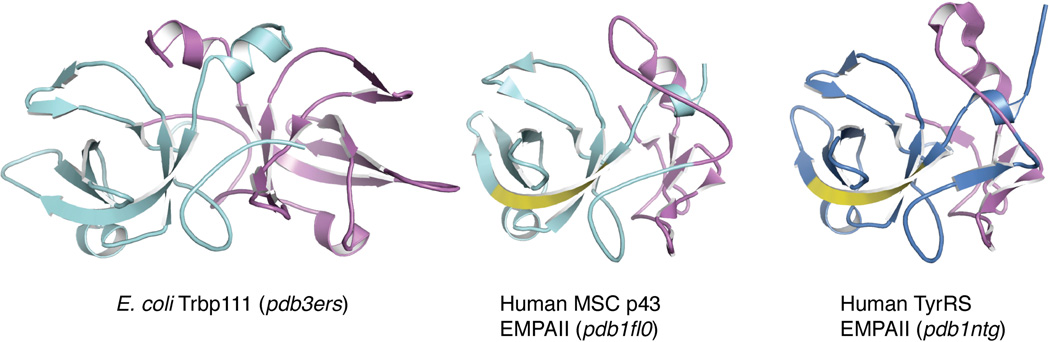
EMAPII domains are only found in aaRSs (C. elegans MetRS and metazoan TyrRS) and in aaRS-associated proteins (p43/AIMP1 and yeast Arc1p (a scaffold protein for the yeast aaRS complex, see below)). This strict distribution of EMAPII domain suggests that its function evolved specifically for tRNA synthetases. The N-terminal portion of EMAP II (∼160 aa) shares sequence homology with Trbp111, a 111 aa, free-standing tRNA binding protein found in bacteria [14]. Structural analysis of EMAPII has revealed that the monomeric EMAPII mimics the dimeric structure of Trbp111 by forming a pseudo dimer interface with its C-terminal sequences (Figure 4) [13]. Trbp111 specifically recognizes tRNA by binding to the elbow region of all tRNAs [15]. In certain bacteria and plants, a Trbp111-like domain is fused to the C-terminus of MetRS to enhance its aminoacylation activity by facilitating the binding of tRNA [16].
Figure 4.
The structure of E. coli Trbp111 and the structures of the EMAPII domain from human MSC p43 and human TyrRS. The potential cytokine motif in EMAPII domains is labeled in yellow.
A domain homologous to EMAPII is present in TyrRS from insects to humans. The EMAP II domain in TyrRS, like many other new domains, is dispensable for aminoacylation. However, removal of EMAPII domain (to generate mini-TyrRS) through natural proteolysis outside the cell activates a cytokine-like function embedded in human TyrRS [17, 18]. Removal of EMAPII appears to expose an otherwise masked tri-peptide ELR cytokine motif in the catalytic domain [19]. A separate cytokine activity associated with the EMAPII domain is also activated when it is released from human TyrRS [17]. Interestingly, the ELR motif is not conserved in bacteria and lower eukaryotic TyrRSs, but only starts to appear from insects, concurrent with the addition of the EMAPII domain.
2.3 GST Domain
The origin of aaRS appended domains is not confined to tRNA-binding motifs. For example, yeast MetRS does not have a Trbp111 domain as seen in the C-terminus of a prokaryotic MetRS (such as E. coli), but instead has a GST domain at its N-terminus. A GST domain sequence is also present in MetRS from insects to humans and in vertebrate ValRS [20–22]. Removal of the GST domain in yeast MetRS abolished the Arc1p–dependent aminoacylation of tRNAMet [23]. Similarly, cleavage of the GST domain in human MetRS rendered the enzyme to be inactive, indicating the importance of the GST domains in aminoacylation [24].
In addition to MetRS and ValRS, two other class I tRNA synthetases, GluRS and CysRS, and three aaRS-associated proteins (the yeast Arc1p, two of the three auxiliary factors of the MSC: MSC p38/AIMP2 and MSC p18/AIMP3) all contain a GST domain [12]. These GST domain-containing proteins are found in complexes with other proteins [25, 26], supporting the general idea that GST domains act as protein-protein binding modules. In fact, GST domains do not necessarily possess enzyme activity and are commonly used for protein assembly and for regulating protein folding (such as the GST domains in the S-crystallins, eukaryotic elongation factors 1-γ (eEF-1γ), 1-β (eEF-1β) and the heat shock protein 26 (HSP26) family of stress-related proteins) [27–29]. Interestingly, the yeast MetRS-Arc1p–GluRS ternary complex is formed through two binary GST-GST domain interactions of MetRS-Acr1p and Arc1p–GluRS (Detailed assembly of the complex will be discussed in the next section) [26]. Consistently, human MetRS was shown to interact with two other GST-domain containing scaffold proteins (p38/AIMP2 and p18/AIMP3) through its GST domain[30–32], and deletion of which abolished the incorporation of MetRS to MSC in cell [16]. On the other hand, although the yeast GluRS is known to interact with Arc1p through their GST domains, the function of the GST domain in human bifunctional GluProRS remains to be defined. The role of the N-terminal GST domain in mammalian CysRS, one of the latest domain additions in aaRS, is also undefined. Intriguingly, two versions of CysRS—one with the GST domain and one without—are produced by alternative splicing in mouse and human [33, 34].
Sequence analysis indicates that all GST domains in aaRSs and in MSC p18/AIMP3 and MSC p38/AIMP2 share strong homology with the eukaryote-specific elongation factors eEF-1β and −1γ [26, 27]. Phylogenetic analysis of these GST domains showed stronger homology with each other than with other common GST proteins or GST domain found in bacteria (Figure 5). Possibly, the GST domains in the aaRS protein family (including the elongation factors) were generated from the same origin by gene duplication events. It is interesting to note that yeast GluRS, MetRS and Arc1p genes are located on the same chromosome (VII). However, how these gene duplication events were specifically confined to aaRS proteins is currently unknown.
Figure 5.
Phylogenetic analysis of GST domains from aaRSs and other GST-containing proteins.
2.4 WHEP Domain
Many appended domains are involved with functions other than aminoacylation, such as the regulations of inflammation, angiogenesis and even p53 activation. The WHEP domain is one example. Initially discovered in three human aaRSs [TrpRS(W), HisRS(H), GluProRS(EP)] , hence its name, the WHEP domain is ∼50 aa long and shares apparent sequence homology among different aaRSs [35]. Two other human aaRSs—GlyRS and MetRS—also contain a WHEP domain at their N- and C-termini, respectively. Early studies, based on sequence similarity and intron positions, suggested that the WHEP domain might have first appeared in HisRS in single-cell eukaryotes (e.g. yeast) and then propagated to other aaRSs [35, 36]. Interestingly, the acquisition of the WHEP domain to GluProRS happened concurrently with the fusion of GluRS and ProRS into one gene, an event that took place prior to the divergence of cnidarians and bilaterians [37]. Among these aaRSs, TrpRS and MetRS appear to have the latest WHEP domain acquisition events that did not occur until the first vertebrates [22, 35]. In contrast to other aaRSs that have only one WHEP domain, GluProRS contains a varying number of WHEP domains depending on the species (3–6). In particular, human GluProRS contains three consecutive WHEP domains in between the N-terminal GluRS and the C-terminal ProRS.
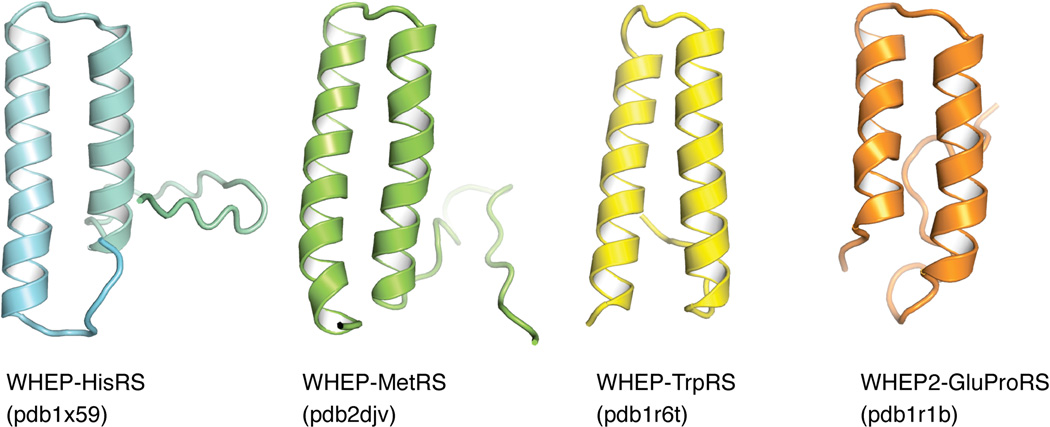
The spreading of this short sequence among several aaRSs suggests that the WHEP domain might be a common tRNA-binding motif [38], though experiments testing this hypothesis have not arrived on a clear conclusion. Structure and functional analyses have indicated that the WHEP domains fold as a simple helix-turn-helix structure and act as a unique RNA recognition motif (Figure 6) [39]. However, the WHEP domain does not significantly affect, at least in in vitro studies, the aminoacylation efficiency or the tRNA binding affinity of their host aaRSs including TrpRS, GlyRS and GluProRS [40–42]. Recent studies revealed that the WHEP domains in human GluProRS perform non-canonical functions through protein-protein and protein-RNA interactions. For instance, the WHEP domains directly mediate the interaction between the synthetase and NSAP1 (NS1-associated protein), L13a and GAPDH (glyceraldeyde 3-phosphate dehydrogenase) to form a gamma-IFN-activated inhibitor of translation (GAIT) complex [43–45], which interacts with eIF4G to block 43S recruitment and mRNA translation [46]. Using their RNA binding property, the WHEP domains are also responsible for recognizing the GAIT element located on the 5’-UTR of target mRNAs [45].
Figure 6.
Structure of WHEP domains in aaRSs.
Although not needed for aminoacylation, the WHEP domain appears to be a regulator for the non-canonical functions of human TrpRS. In human TrpRS, the N-terminal fused single WHEP domain undergoes a conformational change when the synthetase is bound to Trp-AMP (aminoacylation reaction intermediate), which places the WHEP domain close to the active site pocket [47]. The WHEP domain can be specifically removed by proteolysis or alternative splicing to generate fragments of TrpRS (T2-TrpRS and mini-TrpRS, respectively) that exhibit angiostatic activity through its interaction with the extracellular domain of VE-cadherin on the surface of endothelial cells [48–50]. The WHEP domain of TrpRS also mediates direct interactions with DNA-PK (DNA-dependent protein kinase) and PARP-1 (poly(ADP-ribose) polymerase 1) in the nucleus to activate p53 [51]. Finally, it is interesting to note that, like EMAP II domain, WHEP domain only exists within AARS genes. This aaRS-specific domain expansion is suggestive of a special selective pressure to develop new functions for AARS genes during the evolution of higher eukaryotes. Understanding such pressure may be instructive for understanding the various expanded functions of aaRSs, and vice versa.
2.5 Leucine zipper
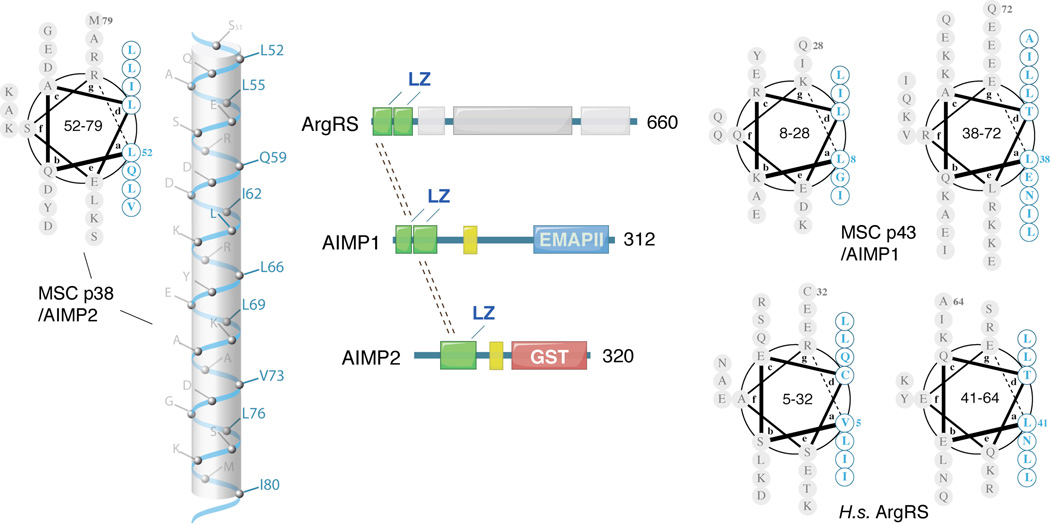
Certain domains expanded in aaRS proteins are only involved in protein-protein interactions. As such, a standard module for protein assembly, the leucine zipper, is often found to mediate protein-protein interactions in many biological processes, such as in forming the snare complex in vesicle trafficking and in forming the trimeric structure of gp41 to facilitate the entry of HIV to target cells [52, 53]. The leucine zipper is a helical motif that has leucine residues (or other hydrophobic residues) at every fourth position of the heptad repeats, so that the protruding isobutyl side chains are lined up on one side of the helix. This design creates a hydrophobic spine that inter-locks with its partner to form a coiled-coil zipper.
Leucine zippers exist in ArgRS (a component of the MSC) in higher eukaryotes from insects to humans, but not in lower eukaryotes such as yeast. Leucine zippers also exist in MSC scaffold proteins p43/AIMP1 and p38/AIMP2, suggesting that they play critical roles in the assembly of the MSC (Figure 7). The leucine zipper motifs in ArgRS interact with the leucine zippers in p43/AIMP1, which in turn interact with the leucine zipper motif in p38/AIMP2 [54]. Therefore, these three proteins may form an ArgRS-p43-p38 subcomplex of MSC through several coiled-coil structures. Interestingly, a shorter form of human cytoplasmic ArgRS, which is produced from an alternative translation initiation site on the same mRNA [55–57], lacks the N-terminal leucine zipper motifs (residue 1–72) and the ability to associate with the MSC.
Figure 7.
Leucine zippers in human ArgRS and aaRS-associated MSC p43/AIMP1 and p38/AIMP2.
Leucine zippers are completely absent from aaRSs that are not associated with the MSC, suggesting that leucine zippers in aaRSs are exclusively used for the assembly of the MSC. A distinct feature of the leucine zipper motif, compared with other protein-protein interaction modules, is its linear and extended geometry, which may be an important characteristic for providing a framework to support the structure of the MCS.
2.6 Unique Domains
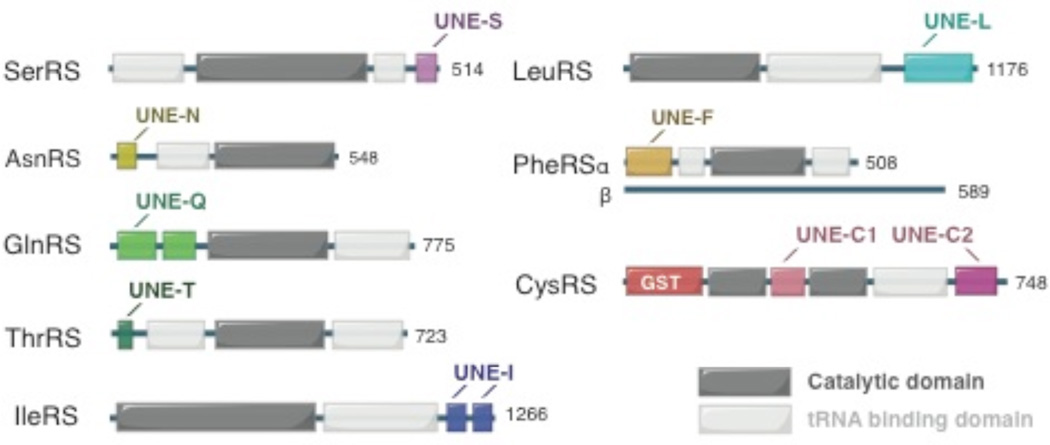
Besides the above mentioned appended domains that are shared among aaRSs or aaRS-associating factors, other domains/sequences that have evolved within eukaryotic aaRSs share no detectable sequence similarity to common structural modules, and for the most part, each domain is present in only one specific aaRS. Because of their uniqueness, these domains are named as UNE-X, where X represents the amino acid-specificity (in single letter code) of the aaRS to which each domain is appended [22].
Recently, several studies have suggested the importance of these unique domains in both canonical and non-canonical functions of aaRSs. Eukaryotic IleRS contains two large additions at the C-terminus (named as UNE-I1 and UNE-I2). UNE-I1 (residue 968–1064) is found throughout eukaryotes (from yeast to human), while UNE-I2 (residue 1065–1266) exists only in vertebrates (Figure 8) [58]. Sequence analysis shows that UNE-I2 contains two repetitive sequences each of ∼90 aa. This region interacts with the WHEP domains of EPRS and therefore may play a role in retaining IleRS in the MSC [31, 59].
Figure 8.
UNE domains as found in human aaRSs.
In higher eukaryotes (from C. elegans to human), LeuRS contains an unique domain at the C-terminus with a size of ∼ 110 aa (Figure 8) [31]. UNE-L is predicted to have rich secondary structures (beta-strands and alpha helices) that presumably fold into a discrete 3D structure. Human LeuRS was recently found to function as a leucine sensor for the mTOR pathway [60–62]. Specially, human LeuRS associates and activates the RagD GTPase of mTORC1 in a leucine-dependent manner. Removal of the C-terminal ∼ 220 residues (including UNE-L and a LeuRS-specific domain) abolished the interaction with RagD [61]. Interestingly, the yeast LeuRS, which does not contain the UNE-L, also controls the TOR pathway. However, in contrast to the human LeuRS, the N-terminal CP1 (editing) domain of yeast LeuRS was proposed to be the binding site for the GTPase, suggesting that the mechanism of LeuRS in regulating the mTOR pathway might be substantially different in yeast as compared to that in mammals [60, 62]. It remains to be determined if the presence of UNE-L has a role in relocating the RagD binding site from the editing domain in yeast to the C-terminus of LeuRS in human.
UNE-F is found at the N-terminus of PheRS α-subunit in eukaryotes (Figure 8). The structure of human PheRS reveals that UNE-F folds into three continuous DNA-binding fold domains (DBD-1, −2, −3) with intervening sequences [63]. Each DBD contains three α helices folded against a three-stranded antiparallel β-sheet. The topology of the DBDs is found in many DNA-binding proteins as well as in double-stranded RNA adenosine deaminase [63]. Modeling of tRNAPhe on to human PheRS suggested that UNE-F interacts with the D, T loops and the anti-codon stem of the tRNA. Deletion of UNE-F abolishes the aminoacylation activity of PheRS, consistent with its predicted role in binding and recognition of tRNAPhe. Interestingly, T. thermophilus PheRS, with a N-terminal structure distinct from the eukaryotic DBD, binds to a specific DNA sequence on its own gene [64]. The presence of three DNA-binding modules in UNE-F suggests that human PheRS might have non-canonical functions involving dsDNA/dsRNA binding such as in transcriptional and translational regulations.
Metazoan and fungal AsnRS differ from their bacterial homologues by the addition of a conserved N-terminal extension of ∼110 aa (UNE-N) (Figure 8). Recent structural characterization showed the UNE-N contains a structured region with a novel fold (residues 1–73) that is connected to the remainder of the enzyme by an unstructured linker (residues 74–110) [65]. Shown by NMR, the folded portion of UNE-N features a lysine-rich helix that interacts with tRNA. Whether UNE-N is also involved with non-canonical functions of AsnRS remains to be determined.
GlnRS is predominately found in eukaryotes, whereas in most prokaryotes, Gln-tRNAGln is synthesized by an indirect pathway to first form Glu-tRNAGln by GluRS, and followed by the conversion of Glu-tRNAGln to Gln-tRNAGln by a tRNA-dependent amidotransferase. Compared to the few existing bacterial GlnRSs, the eukaryotic enzymes contain a N-terminal extension of ∼200 aa (UNE-Q) (Figure 8). Yeast mutants lacking UNE-Q exhibit growth defects and have reduced complementarity for tRNAGln and glutamine [66]. Structural analysis shows that UNE-Q consists of two subdomains that resemble the two adjacent tRNA specificity-determining domains in the GatB subunit of GatCAB, the trimeric amidotransferase that can use both Glu-tRNAGln and Asp-tRNAAsn as substrates to form Gln-tRNAGln and Asn-tRNAAsn, respectively. The two subdomains of UNE-Q are connected by a conserved hinge region, which when mutated, reduced yeast GlnRS’ affinity for tRNAGln. UNE-Q gives another example that the domain addition or exchange is highly selective for aaRSs or aaRS-related genes.
In addition to the N-terminal GST domain that seems to only exist in mammals, all eukaryotic CysRS contains two other sequence additions (UNE-C1 and UNE-C2) (Figure 8). UNE-C1 is inserted in human CysRS between residues 108 and 223, and in front of the well-known CP1 insertion (residue 273–419). UNE-C2 is a C-terminal extension of ∼150 aa. Both UNE-C1 and UNE-C2 are unique to CysRS, and show no apparent sequence homolog to other domains or motifs.
The smallest UNE domain is the UNE-S motif (∼30–40 aa) that is located at the C-terminus of vertebrate SerRS (Figure 8). UNE-S was found to be essential for vascular development [67]. Deletion of the C-terminal sequences of SerRS, including UNE-S, led to abnormal blood vessel formation that resulted in premature death in zebrafish [68, 69]. Removal of UNE-S has little effect on the aminoacylation activity of human SerRS. Further studies discovered a significant portion of SerRS is in the nucleus of human umbilical vein endothelial cells, and the localization is directed by a classical nuclear localization signal (NLS) embedded in the UNE-S. Interestingly, SerRS regulates the expression of VEGFA in the nucleus through an unknown mechanism. This novel function is independent of the canonical aminoacylation activity of SerRS, as an aminoacylation-defective form of full length SerRS could fully rescue the vascular phenotype in zebrafish [68]. Therefore, acquisition of UNE-S appears to be important for the non-canonical function of vertebrate SerRS.
In summary, higher eukaryotic aaRSs continue to evolve with the additions of new domain/sequences. Many of these domains are related to their canonical function, especially their tRNA binding properties, while others are dispensable for aminoacylation. Although the timing of each domain addition may be different, one common feature is that once a domain is added to an aaRS, the process is generally irreversible and the domain is conserved from then on as an integral part of that aaRS. Therefore, the timing of domain acquisition may be linked to the new biology associated with increasing complexity of the organism and could provide important hints for their potential functions beyond the canonical aminoacylation function of their prokaryotic homologues [22]. We predict that more non-canonical functions will be discovered that link to the acquisitions of the UNE domains and other appended domains of aaRSs.
3 Emergence of Multi-Synthetase Complex in Higher Eukaryotes
Prokaryotic aaRSs are a large family of ∼ 20 proteins that perform a common function, namely, aminoacylation of tRNA Unlike many other housekeeping machineries, such as the RNA polymerase complex and the ribosome, they do not form a complex. This feature is presumably related to the independence of the aminoacylation activity for each amino acid and their tRNAs. It might even be beneficial to not have aaRSs too near each other in a complex, as they might otherwise sterically hinder each other for tRNA-binding or attract tRNA nonspecifically, which could increase the chance of mis-binding and mis-charging. Although several small (binary/ternary) aaRS complexes have been found in archaea, they appear to occur serendipitously in only certain species, rather than being a common feature of the family [70, 71]. Thus the emergence of such a conserved, stable, and large complex of aaRSs in higher eukaryotes is remarkable.
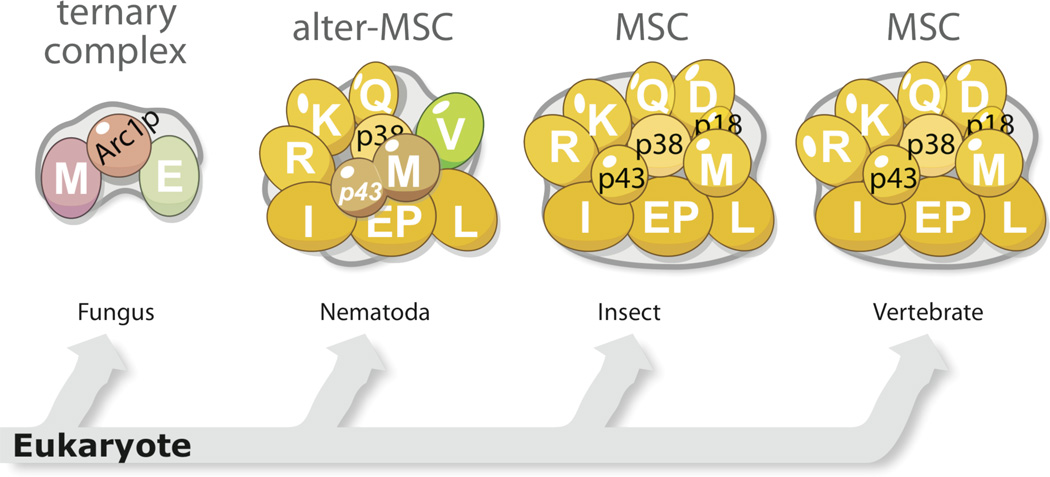
3.1 A miniature complex in yeast
In eukaryotes, the complexity of an aaRS complex increases with the complexity of the hosting organism. For instance, in the single celled yeast (S. cerevisiae), the aaRS complex is a relatively simple ternary complex comprising MetRS, GluRS and Arc1p (aminoacyl-tRNA synthetase cofactor 1 protein) (Figure 9) [72]. With a GST domain at the N-terminus and a EMAPII-like domain at the C-terminus, Arc1p is essentially a fusion protein that links a protein-binding module to a tRNA-binding domain [73]. Concurrently, both MetRS and GluRS in yeast acquired an N-terminal GST domain. It is through the GST domains that Arc1p links MetRS and GluRS together [74].
Figure 9.
Evolution of aaRS complexes in eukaryotes.
As mentioned in 2.2, Trbp111 is a non-specific tRNA-binding domain. The Trbp111-like domain in Acr1p not only facilitates the binding of tRNAMet and tRNAGlu to the complex, but also nonspecifically binds to other tRNAs [23]. Although Arc1p is not essential in vivo, it enhances the aminoacylation activity by two orders of magnitude for MetRS and by one order of magnitude for GluRS as shown by in vitro kinetic studies [75]. It is worth noting that the E. coli MetRS has a Trbp111 domain at its C-terminus. Therefore, the same strategy for enhancing MetRS activity in E. coli is also used in yeast through the formation of a MetRS-Arc1p–GluRS complex [73]. Interestingly, a recent study reported the MetRS-Arc1p–GluRS complex can bind and mismethionylates many tRNA species in vitro [76]. Moreover, a similar effect on tRNA mismethionylation was achieved by fusing the Trbp111 domain of Arc1p to the yeast MetRS [77].
The MetRS-Arc1p–GluRS complex has a number of other functions. One is to regulate the cytoplasmic localization of MetRS and GluRS. Disruption of the complex by deletion of the GST-domain of Arc1p resulted in strong nuclear localization of all three components [78]; GluRS and Arc1p were found to associate with apurinic/apyrimidinic sites of damaged DNA in the nucleus [79]. In addition, the MetRS-Arc1p–GluRS complex is also important for GluRS trafficking into the mitochondria, which is important for providing an alternative pathway to the GlnRS activity that is lacking in yeast mitochondria [80, 81]. In most bacteria, archaea and organelles of many eukaryotes, including yeast mitochondria, Gln-tRNAGln is generated through an indirect pathway, whereby a nonspecific GluRS synthesized Glu-tRNAGln, and then an aminotransferase converts it to Gln-tRNAGln. However, the yeast mitochondrial GluRS cannot mischarge tRNAGln [82]. Instead, the cytoplasmic GluRS is transported to the mitochondria to charge the mitochondrial tRNAGln with glutamate. The cellular level of Arc1p controls the translocation of GluRS. Interestingly, the expression of Arc1p is downregulated when yeast switches from fermentation to respiratory metabolism, which comes with a high demand for mitochondrial protein synthesis [80].
In addition, Arc1p is potentially involved with tRNA maturation and exportation from the nucleus. Deletion of Arc1p showed synthetic lethality when combined with the deletion of tRNA transporter Los1, which is responsible for transporting mature tRNAs from the nucleus to the cytosol [23, 83]. Overall, formation of this prototypic aaRS complex is to anchor GluRS and MetRS in the cytosol and to facilitate the aminoacylation function of the two synthetases by providing them with an additional tRNA-binding module – similar roles are found in further developed complexes in higher species.
3.2 A divergent complex in C. elegans
In ascending the tree of life, eukaryotes evolved from single-celled to multicellular organisms, which allows for the differentiation of cells to have specialized roles. Compared to fungi, the sizes of aaRSs in C. elegans are largely increased by the addition of appended domains. Extensive interactions between new domains present in C. elegans, but not in fungi, appear to facilitate the formation of the aaRS complex in C. elegans, which shares 7 aaRS components with the most highly evolved mammalian MSC. A “pull down” assay revealed that MetRS associates with a complex that contains a MSC p38/AIMP2 homologue and 8 aaRSs including LeuRS, IleRS, GluRS, ValRS, MetRS, GlnRS, ArgRS and LysRS (Figure 9) [84]. The only exception is ValRS, which is found in the C. elegans complex but not in the mammalian MSC. No appended domain is found in C. elegans ValRS, and what enables ValRS to be associated with the C. elegans aaRS complex remains unclear. On the other hand, 2 aaRS components of the modern MSC (AspRS and ProRS) and 2 accessary factors (p43/AIMP1 and p18/AIMP3) are missing in the C. elegans complex. Interestingly, C. elegans MetRS has a C-terminal extension (∼320 aa) that contains a leucine zipper and an EMAPII domain that shares extensive sequence homology with mammalian MSC p43/AIMP1. Functional studies have confirmed that human p43 can substitute the C-terminal extension of C. elegans MetRS for its in vivo activity [85]. Therefore, C. elegans MetRS can be viewed as a combination of MetRS and p43/AIMP1.
Although the C. elegans aaRS complex and the modern MSC share a significant degree of homology, some evidence suggests that not all interactions within the complexes have arisen through convergent evolution. Recent phylogenetic analysis of the GluRS and ProRS genes showed that the fusion of the two genes into one bifunctional GluProRS gene appeared before the Bilateria, and that a fission event happened in C. elegans that separated the EPRS gene back to the two genes [37]. This analysis supported the idea that the distinct C. elegans aaRS complex is, at least with respect to some of its components, the result of divergent evolution [84].
3.3 Multi- tRNA synthetase complex from fruit fly to human
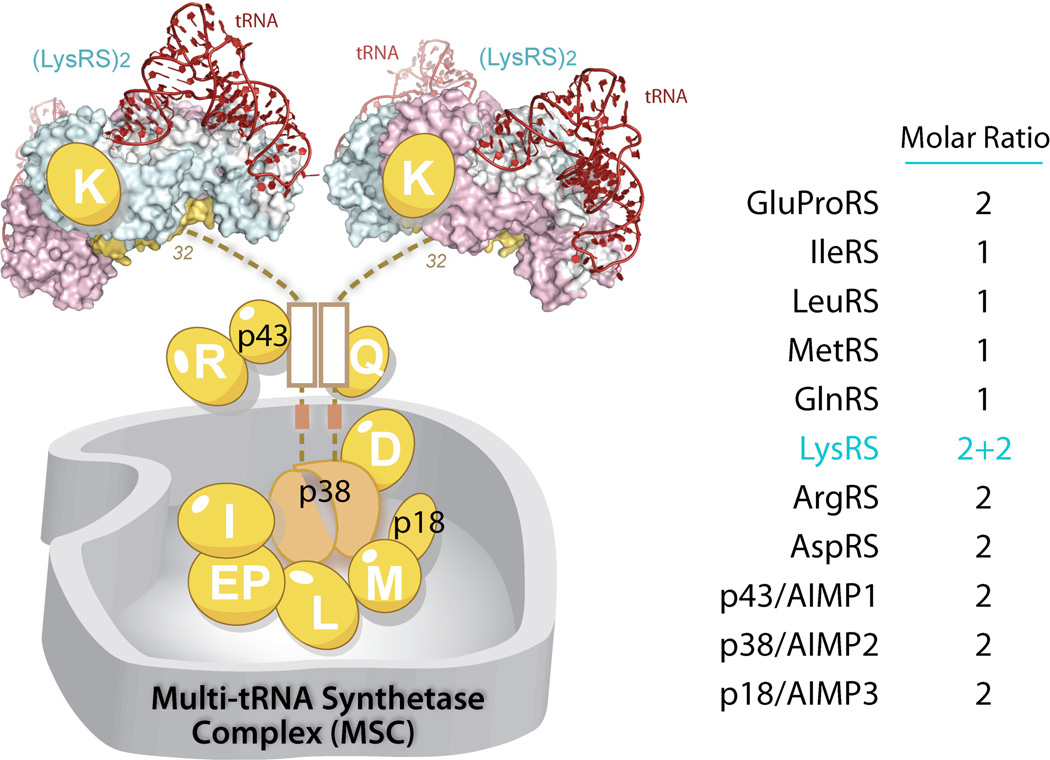
It has long been proposed that the MSC is a ubiquitous cellular component of higher eukaryotes [86]. Its presence has been documented in various higher eukaryotes including rabbits, sheep, bovines, mice, rats and humans, as well as in flies (Drosophila), and genes encoding the three MSC scaffold proteins—p43, p38, p18—are found in all representative species from insects to humans [22]. Also named as aminoacyl-tRNA synthetase interacting multi-functional protein 1, 2, 3 (AIMP1, 2, 3), these 3 scaffold proteins interact with each other and with the 9 AARS components (LysRS, ArgRS, GlnRS, AspRS, MetRS, IleRS, LeuRS and GluProRS) to promote assembly of the MSC with a molecular weight of 1.5 million Da (Figure 9). The function(s) of MSC is unclear but has been proposed to facilitate channeling of aminoacylated tRNA to the ribosome during protein synthesis [87], and to provide a cellular reservoir of various non-canonical functions of its synthetase and non-synthetase components, in response to certain stimulation or environmental cues [25, 88].
The stoicheometry of the MSC components remains largely consistent, from Drosophila to human [86, 89] (Figure 10). However, the stoicheometry appears to vary upon conditions in cell. For example, one study reported that the number of MSC p43/AIMP1 increased from 2 to 4 molecules per MSC, when the protein was overexpressed in the cell [90]. Also, when the cellular level of methionine decreased, the amount of MetRS in the MSC was observed to double to 2 molecules per MSC [91].
Figure 10.
Multi-aminoacyl-tRNA synthetase complex (MSC) in higher eukaryotes.
A number of studies suggest that whereas the MSC serves as a reservoir for almost half of the cellular tRNA synthetases, it also controls the flow of synthetases between their canonical and noncanonical functions [88, 92]. As evidence for the latter function, many MSC components have been reported to be released from the complex under certain conditions. For example, LysRS is released upon antigen-IgE induced mast cell activation [93]; EPRS is released for the resolution of INF-γ related inflammation [43]; GlnRS is released during Fas-ligand triggered apoptosis. Interestingly, even the scaffold proteins can be released [94]. MSC p43/AIMP1 is released and secreted to act as a cytokine under stress conditions [95]; MSC p38/AIMP2 is released upon DNA damage and translocated to the nucleus to activate p53, FBP and TRAF2 [96–98]. MSC p18/AIMP3 is released under UV radiation and is also translocated to the nucleus [99, 100].
How MSC is assembled through various interactions, and how the assembly would allow for specific release of its individual components have been interesting topics for more than two decades. Early studies employing gene knockdowns of each component and by crosslinking approaches have mapped out the interactions within the MSC [30, 31, 101–103]. Interestingly, for understanding the evolution of this complex, certain interactions found in the early yeast complex are conserved in the MSC. For example, the interactions between the GST domains of MetRS, Arc1p and GluRS in the yeast complex are likely to be maintained in the MSC between the GST domains of MetRS, p38 and p18 as revealed by crystal structure analysis of human MSC p18/AIMP3 [74]. On the other hand, the MSC p43/AIMP1 and p38/AIMP2 are proteins newly invented in higher eukaryotes, and are responsible for a number of interactions specific to the MSC.
The MSC component p38/AIMP2 is the core of the MSC, and directly interacts with most of the MSC components, including LysRS, GlnRS, AspRS, GluProRS, and IleRS as well as p43 and p18 [30, 90, 101, 104]. Depletion of MSC p38 leads to complete disruption of MSC [30, 105]. Mapping studies indicated that p38/AIMP2 interacts with other MSC components in a linear and sequence-dependent manner. The N-terminal end of p38/AIMP2 interacts with LysRS; the following coiled-coil region interacts with p43/AIMP1 and ArgRS; a less structured linker interacts with GlnRS; and finally, the C-terminal GST domain binds to p18/AIMP3, AspRS, MetRS and other aaRSs (Figure 10).
Among those interactions, the LysRS and p38 interaction appears to be most critical for the stability of MSC. Sequence alignment shows that the LysRS interacting residues in p38 are highly conserved, supporting the idea that the MSC assembly, especially the LysRS:p38/AIMP2 interaction, is conserved in all higher eukaryotes [106]. In functional tests, gene knock-downs of LysRS led to MSC disruption and subsequent degradation of MSC components [105]. A further study, using recombinant proteins, reconstituted the subcomplex of LysRS:p38 in vitro. Surprisingly, the LysRS:p38 subcomplex showed a (2:1)x2 stoicheometry with each p38 subunit binding to one LysRS dimer through its N-terminal sequences [107]. Because p38 forms this dimer through its C-terminal GST domain, this explains the long observed stoichiometry of 4 subunits of LysRS present in the MSC, the highest among all MSC components [89]. Presumably, this arrangement of mutually independent bindings of two LysRS dimers for p38 allows for specific release of one LysRS dimer under certain response, while maintaining the stability of the MSC with the other LysRS dimer [93]. This novel stoichiometry was later confirmed by a high resolution co-crystal structure of human LysRS-p38, the first subcomplex structure of MSC. The structure shows that the N-terminal 32 residues of p38 are composed of two short motifs, which sequentially bind to two symmetric grooves of the dimeric LysRS [106].
4 Metamorphosis of tRNA synthetases
Structural metamorphosis of aaRSs has occurred during evolution by the addition of new domains, and has played a critical role in the activation and regulation of the expanded functions of aaRSs. Different functions of an aaRS may be represented by distinct conformations of the same aaRS, and each aaRS may be viewed as a collection of various conformational states that can be converted from one to the other. The various conformations of aaRS provide the structural basis for obtaining different interaction partners in different cellular or extracellular milieus that dictate its functionality, and provide ways of regulation in response to environmental stimulus or developmental cues.
The conformational change of an aaRS protein can be achieved by various ways. One mechanism is through proteolysis to remove part of the protein sequence, thus to expose new areas that were masked or hindered in the conformation of the full-length protein. Such conformational resection can also be achieved by alternative splicing at the mRNA level, with subsequent translation of the splice variants into truncated protein sequences. In addition, alternative splicing-based changes are, in principle, capable of creating bona fide structures. Other mechanisms to generate alternative conformations without a resection are exemplified as posttranslational modifications or disease-associated missense mutations, which have been identified in the human population. A conformational change in protein may further trigger a shift in its structural organization at the quaternary level.
4.1 Proteolysis-based structural resection
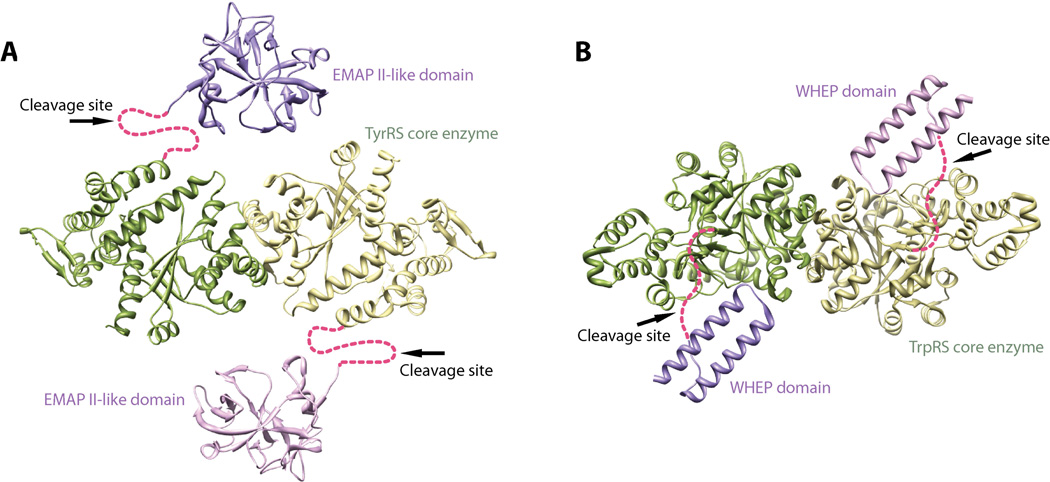
Proteolysis has been observed with several human aaRSs at specific regions. Those regions are usually linkers that join the evolutionary conserved aaRS enzyme core with a new domain incorporated at a later stage in evolution. Interestingly, these linkers are usually disordered in crystal structures of aaRSs or, for those without crystal structure information, are predicted to be unstructured. High flexibility of the linkers may render them to be more accessible for various proteases. Interestingly, proteolysis that removes an appended domain from the core enzyme has been shown to be associated with activation of a non-canonical function of aaRS. For example, the EMAP-II-like domain appended to human TyrRS is linked to the core enzyme (named as mini-TyrRS) via an unstructured loop of 22 amino acids (D343-I364) that is disordered in the crystal structure [108]. This loop is cleaved by at least two proteases—plasmin and leukocyte elastase—with different sequence specificities (Figure 11A) [17, 109]. The cleavage at this loop activates the cytokine activities of both mini-TyrRS and EMAP-II, which are otherwise mutually inhibited in the context of the full-length protein. Another example is human TrpRS, which is appended with a helix-turn-helix WHEP domain. The WHEP domain is linked to the core enzyme of TrpRS via a 29-residue loop (G55-D83) in which 21 residues (D61-E81) were disordered in the crystal structure. Similar to TyrRS, the loop contains the cleavage site for both plasmin and leukocyte elastase [49, 110], which cleave off the WHEP domain from the core enzyme to activate its angiostatic activity (Figure 11B).
Figure 11.
Appended domains join to TyrRS (A) and TrpRS (B) enzyme cores via flexible linkers that serve as cleavage sites for the activation of novel functions of TyrRS and TrpRS.
Addition or removal of an appended domain might not dramatically affect the conformation of the core enzyme. This scenario has been demonstrated by human TrpRS, where the core enzyme adapts essentially the same conformation as a stand-alone protein and as part of the full-length protein [111]. However, as a result of an appended domain being removed, the overall conformation of an aaRS must be affected and certain areas may become exposed. In case of human TyrRS, removal of the EMAP II domain exposes the ELR motif that is critical for mediating the angiogenic and cytokine-like activity of mini-TyrRS [17]. As for human TrpRS, removal of the WHEP domain opens the active site that has both a Trp and an ATP binding pocket. The active site is used to bind to two Trp residues near the N-terminus of VE-cadherin, and the binding blocks the hemophilic interaction between VE-cadherins that is critical for angiogenesis [48–50]. Importantly, a flexible and long linker region, like that found in TyrRS and TrpRS, would allow for the incorporation of more than one protease recognition site to facilitate cleavage at the linker region.
4.2 Alternative splicing-based structural resection
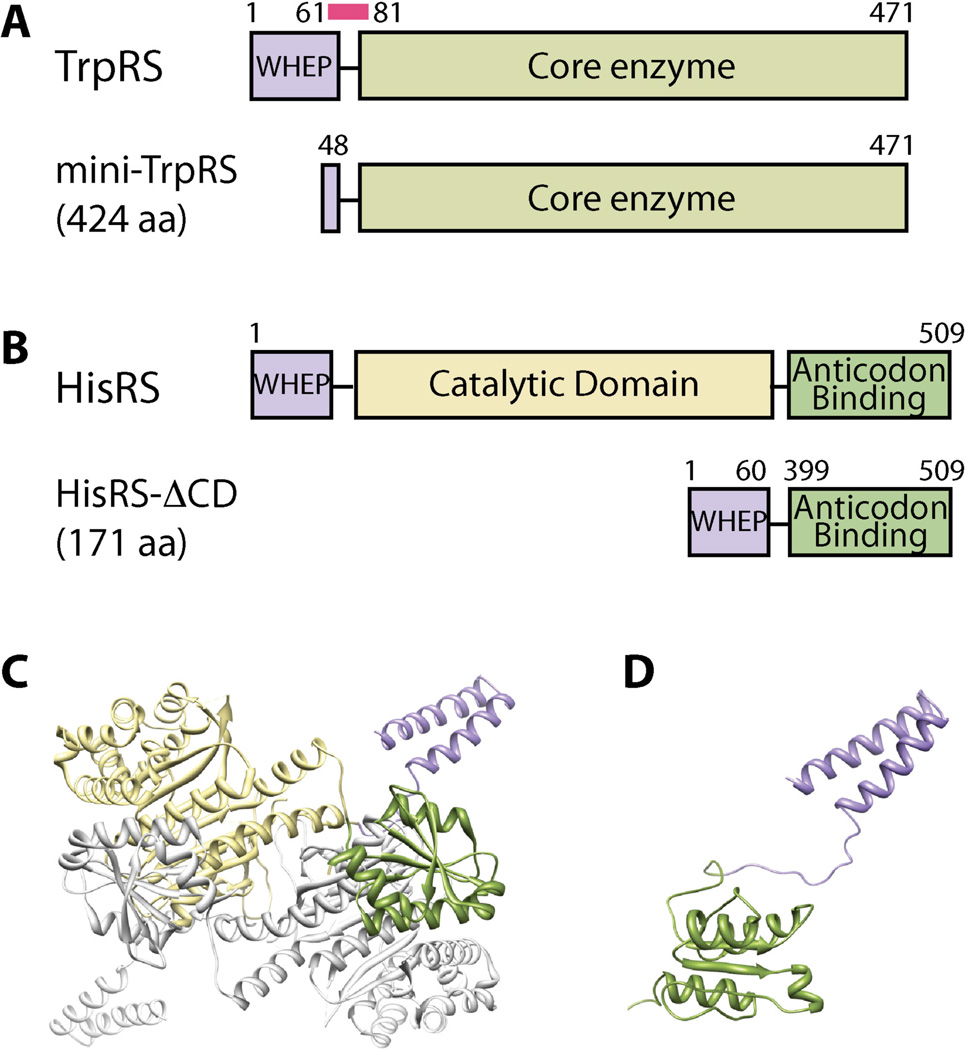
As mentioned above, alternative splicing at the mRNA level can also achieve structural resection of aaRSs. For example, an exon-skipping event generates an mRNA variant of human TrpRS that lacks exon 2. The splice variant is translated into a shorter form of TrpRS (mini-TrpRS) that missing the first 47 residues and the majority of the WHEP domain [112–114]. Mini-TrpRS, similar to the other WHEP domain-deleted forms of TrpRS generated by proteolysis, exhibits angiostatic activity that is masked in the full-length protein (Figure 12A) [115].
Figure 12.
Alternative splicing generates structural resections of aaRSs. A. Human TrpRS and its splicing isoform; B. Human HisRS and its splicing isoform; C–D. Structure of human full-length HisRS (C) and the alternatively spliced isoform (D).
Alternative splicing can also achieve internal deletion, which is not possible by proteolysis. A recent example is an exon-skipping event on human HisRS that removes a large segment of mRNA from exon 3 to exon 10. This event results in the precise deletion of the entire catalytic domain (CD) to make a protein product that directly links the N-terminal WHEP domain to the C-terminal anticodon-binding domain [116]. NMR spectroscopy analysis revealed a dumbbell-like conformation of the splice variant with the WHEP and anticodon-binding domains loosely linked together. Although the conformation of each domain is more or less preserved, the overall tertiary and quaternary structures of the splice variant dramatically differ from that of the full-length HisRS (Figures 12B–D).
In principle, as alternatively spliced mRNAs are translated into new polypeptides, they can generate independent new structures. The new conformations could result from new protein sequences that are being created, for example, by intron retention or frame shift events, or from sequence deletions that may or may not affect the conformation of the rest of the proteins. In case an internal deletion takes place in the middle of a globular domain (rather than at the domain boundary, as in the HisRS case above), a bona fide new structure may be generated that would not be possible to generate by proteolysis.
4.3 Posttranslational modification–based structural change
Posttranslational modifications such as phosphorylation have also been found to control the non-canonical functions of aaRS proteins,, presumably through posttranslational modification–based conformational change. A well-studied example is EPRS, the dual functional tRNA synthetase comprised of a N-terminal GluRS and C-terminal ProRS that are linked together through three consecutive WHEP domains. EPRS is a component of the MSC. Upon interferon-γ stimulation, EPRS is phosphorylated at Ser886 (in between the second and the third WHEP domains) and at Ser999 (after the third WHEP domain and before the C-terminal ProRS), and both phosphorylation events are required to trigger the release EPRS from the MSC [44]. Although the structural change is undefined, phosphorylated EPRS, but not its unphosphorylated form, can facilitate the assembly of a heterotetrameric γ-interferon-activated inhibitor of translation (GAIT) complex that binds to mRNAs with GAIT elements to suppress their translation.
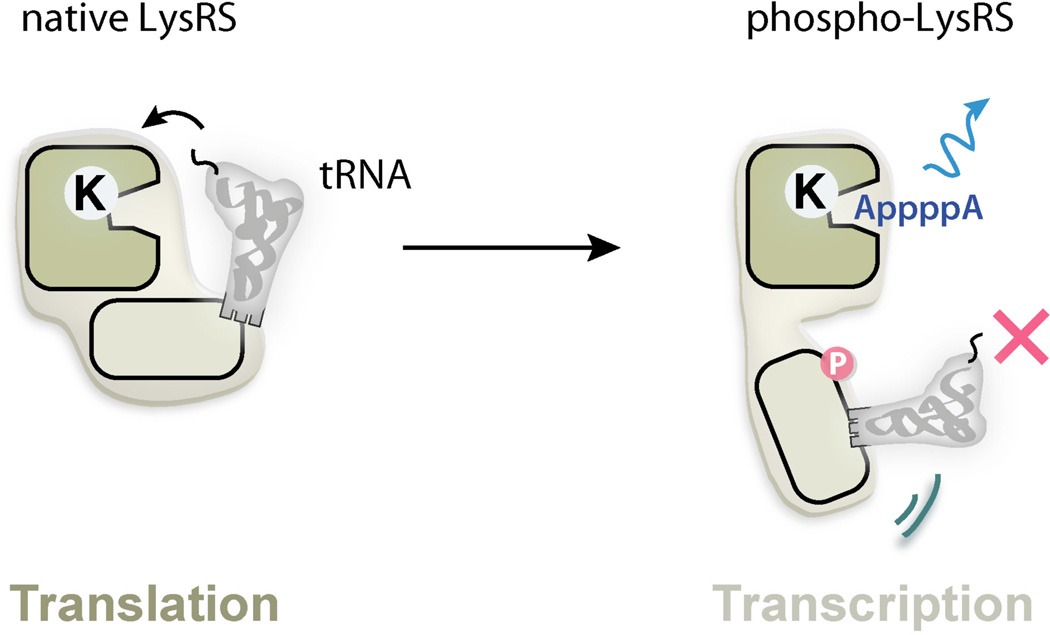
Another example is LysRS. Upon immunological challenge, LysRS in mast cells is phosphorylated at Ser207, which triggers the release of LysRS from the MSC and its subsequent nuclear localization to regulate transcription factor MITF [93]. In the absence of Ser207 phosphorylation, LysRS is strongly associated within the MSC in a “closed” form to catalyze the aminoacylation reaction that charges lysine onto tRNALys for protein synthesis [106]. However, phosphorylation at Ser207 triggers a distinct conformational change that opens up the structure. As a result, phosphorylated LysRS is released from the MSC and translocated from cytoplasm to the nucleus, where it binds to MITF, and generates diadenosine tetraphosphate (Ap4A) to activate the transcription of MITF target genes. The open conformation can no longer aminoacylate tRNA but has significantly elevated activity in Ap4A production [106]. Therefore, phosphorylation-based conformational change switches the function of LysRS from translation to transcription (Figure 13). Significantly, phosphorylation at a different site, Thr52, translocates LysRS to cell membrane, where it interacts with the 67LR laminin receptor and enhances laminin-dependent cell migration in breast cancer cells [8]. Therefore, the observations that two phosphorylation events on one LysRS protein lead to two completely different signaling cascades further indicates the high potential of aaRS structural metamorphosis in regulating the cellular pathways [8].
Figure 13.
Post-translational modifications generate structural metamorphosis of LysRS.
In addition to phosphorylation, other types of posttranslational modifications such as acetylation and neddylation have been found on aaRS [117–119]. Although not yet characterized, the modifications could also induce structural and functional changes on their target aaRSs.
4.4 Mutation-induced structural change
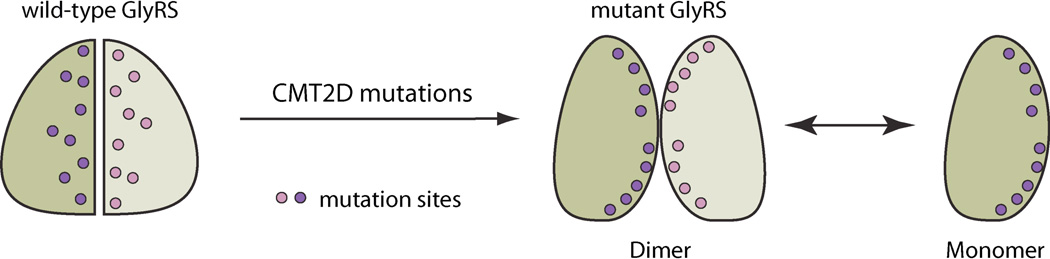
Mutations in aaRS genes have been associated with various diseases, and most prominently with a genetic disorder named Charcot-Marie-Tooth (CMT) disease. CMT disease is the most common heritable peripheral neuropathy affecting approximately 1 in 2,500 people [120]. More than forty genes have so far been associated with CMT through mutations that lead to similar clinical presentations that are characterized by loss of muscle tissue and touch sensation in body extremities [121]. Among them, four genes encode aaRSs (i.e. GARS, YARS, AARS and KARS, encoding GlyRS, TyrRS, AlaRS and LysRS, respectively) and thus make aaRS one of the largest gene families associated with CMT.
Among the four aaRSs linked to CMT, GARS mutations were most frequently found in CMT patients. Eleven mutations in GARS have been linked to an axonal type of CMT (CMT2D), and two separate mutations were found in mice to cause CMT2D–like phenotypes. The mutations are distributed throughout the protein in multiple domains, and do not always affect the enzymatic activity of the synthetase [122]. A gain-of-function mechanism has been clearly demonstrated using the mouse model Nmf249, where the CMT2D–like phenotype that is linked to a spontaneous mutation P234KY cannot be rescued by expression of WT GlyRS [123].
Crystal structure analysis did not reveal significant conformational change caused by a CMT2D mutation, presumably because the potential conformational change is restrained by the crystal lattice interactions [124]. However, a study using a solution method (i.e. hydrogen-deuterium exchange analysis), found that different CMT2D–causing mutations induce similar and dramatic conformational change that opens up the structure to expose a consensus area that is otherwise masked in the WT protein [125]. Based on this study, mutations in this consensus area are hypothesized to be responsible for mediating a gain-of-function interaction that leads to pathological sequelae found in CMT2D patients (Figure 14).
Figure 14.
CMT2D-causing mutations generate structural metamorphosis of GlyRS.
Although a gain-of-function mechanism has not yet been demonstrated with other aaRS proteins linked to CMT, it is possible that a similar mutation-induced structural change may be involved. This consideration is based on the fact that almost all aaRS-associated CMT mutations are dominant, consistent with gain-of-function mutations [126–129]. In addition, CMT-causing mutations are predominantly located near subunit or domain interface, a presumably sensitive location to trigger conformational change. For example, all 13 CMT2D mutation-linked residues in GlyRS are located near the dimerization interface and approximately half of them make direct dimer interactions [122]. Similarly, CMT-linked residues in LysRS are distributed at the dimer interface [130, 131].
4.5 Quaternary structural change
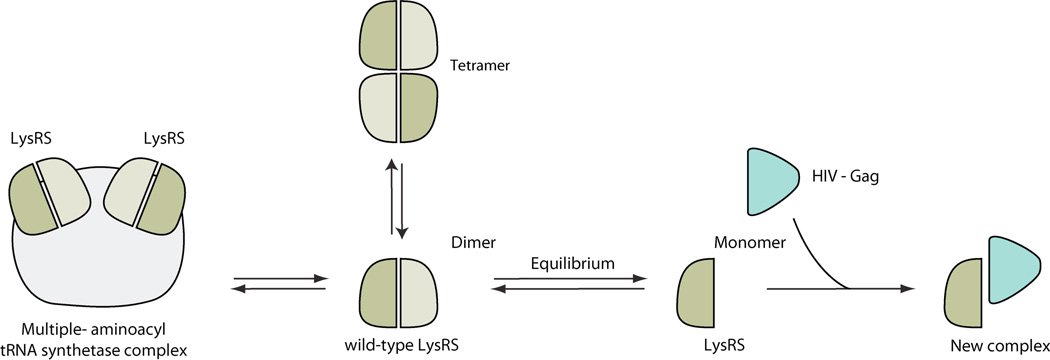
About half of the aaRS family members form a quaternary structure (dimer, tetramer, or heterodimer) in order to be catalytically active for aminoacylation. The most common form of a quaternary structure for aaRSs is a dimer. Interestingly, at least for some aaRSs, the dimer interface is considerately smaller in the human proteins as compared to their bacteria counterparts [111]. Presumably, the reduced dimer interface corresponds to a higher propensity for subunit dissociation, whereby the resulting monomeric conformation may be associated with functions that are distinct from the dimer form. An example is human TyrRS. Although both the monomer and the dimer form of mini-TyrRS can bind to the cell surface receptor CXCR1/2, only the monomer form can induce the migration of PMN cells [132]. Another example is LysRS. The monomeric form of LysRS is suggested to interact with the capsid Gag protein of HIV, which helps the packing of tRNALys, a primer for viral reverse transcription, into the HIV virion [133]. Interestingly, although monomeric LysRS is inactive for aminoacylation [134], disruption of LysRS dimerization seems to not have a major affect on tRNA binding [135]. These examples raised the possibility that monomer-dimer equilibrium is a mechanism to regulate the aminoacylation (dimer) and the novel (monomer) functions of aaRSs.
The catalytically active dimer form not only can dissociate into monomers, but also can further associate to form tetramers (Figure 15). For example, purified human LysRS is found in solution to exist as dimers and as tetramers [107, 130]. Inside the cell, LysRS is a component of the MSC through its interaction with AIMP2/p38, a scaffold protein required for the assembly of the MSC. The binding surface for AIMP2/p38 on LysRS overlaps with the dimer-dimer interface of the LysRS tetramer [107], therefore if such tetramers exist in vivo, they may regulate the assembly of the MSC.
Figure 15.
Structural metamorphosis of aaRSs generated by quaternary structure changes.
A conformational change at the quaternary level can also be combined with, or regulated by, conformational changes caused by other mechanisms. For example, the aforementioned splice variant of HisRS can no longer form dimers as the dimer interface is embedded in the catalytic domain which is ablated in the splice variant [116]. Interestingly, the conformational opening of GlyRS by CMT2D–causing mutations alters the dimerization interface, and either inhibits or promotes dimer formation, depending on the mutation [125].
5 Concluding Remarks
The number of genes that can be traced back to form the genetic core of common ancestor is extremely small (∼80) [136]. More than half of them are translation machineries, including tRNA synthetases, ribosomal proteins and their related factors. These are the only members in the rest of the proteome that match the aaRS family from the point of evolution. When compared to these conserved protein families, such as ribosome proteins, aaRS proteins have shown a high propensity to add new sequences and especially, new domains during the evolution of higher eukaryotes. Why is the expansion of new domains/sequences is unique among the fundamental protein machinery, but universal to the aaRS family? And why are aaRS proteins particularly prominent in their capacities to mediate a broad range of functions beyond translation? These fundamentally intriguing questions remain unanswered. The uniqueness of aaRSs may be connected to some of their special features, such as their long exposure to evolutionary processes, their ubiquitous nature and presence in all life forms, their modular domain architectures, their diverse array of specific amino acid binding pockets, or their unique ability to coordinate translation with other cellular processes. Possibly, the answer lies in the combination of all the features above, and more to be discovered in the future.
Contributor Information
Min Guo, Department of Cancer Biology, The Scripps Research Institute, 130 Scripps Way, Jupiter, FL 33410, USA, GuoMin@scripps.edu.
Xiang-Lei Yang, Department of Cancer Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037, USA, xlyang@scripps.edu.
References
- 1.Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Mirande M. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- 3.Cheong HK, et al. Structure of the N-terminal extension of human aspartyl-tRNA synthetase: implications for its biological function. Int J Biochem Cell Biol. 2003;35(11):1548–1557. doi: 10.1016/s1357-2725(03)00070-0. [DOI] [PubMed] [Google Scholar]
- 4.Liu S, et al. (1)H, (13)C and (15)N resonance assignment of the N-terminal domain of human lysyl aminoacyl tRNA synthetase. Biomol NMR Assign. 2012 doi: 10.1007/s12104-012-9430-x. [DOI] [PubMed] [Google Scholar]
- 5.Francin M, Mirande M. Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase. J Biol Chem. 2003;278(3):1472–1479. doi: 10.1074/jbc.M208802200. [DOI] [PubMed] [Google Scholar]
- 6.Francin M, et al. The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J Biol Chem. 2002;277(3):1762–1769. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- 7.Cen S, et al. Ability of wild-type and mutant lysyl-tRNA synthetase to facilitate tRNA(Lys) incorporation into human immunodeficiency virus type 1. J Virol. 2004;78(3):1595–1601. doi: 10.1128/JVI.78.3.1595-1601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DG, et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 2012 doi: 10.1096/fj.12-207639. [DOI] [PubMed] [Google Scholar]
- 9.Frugier M, Moulinier L, Giege R. A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 2000;19(10):2371–2380. doi: 10.1093/emboj/19.10.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escalante C, Yang DC. Expression of human aspartyl-tRNA synthetase in Escherichia coli. Functional analysis of the N-terminal putative amphiphilic helix. J Biol Chem. 1993;268(8):6014–6023. [PubMed] [Google Scholar]
- 11.Jacobo-Molina A, Peterson R, Yang DC. cDNA sequence, predicted primary structure, and evolving amphiphilic helix of human aspartyl-tRNA synthetase. J Biol Chem. 1989;264(28):16608–16612. [PubMed] [Google Scholar]
- 12.Guo M, Schimmel p, yang xL. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584(2):434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renault L, et al. Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry. EMBO J. 2001;20(3):570–578. doi: 10.1093/emboj/20.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales AJ, Swairjo MA, Schimmel P. Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO Journal. 1999;18(12):3475–3483. doi: 10.1093/emboj/18.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomanbhoy T, et al. Simultaneous binding of two proteins to opposite sides of a single transfer RNA. Nat Struct Biol. 2001;8(4):344–348. doi: 10.1038/86228. [DOI] [PubMed] [Google Scholar]
- 16.Kaminska M, et al. A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J. 2000;19(24):6908–6917. doi: 10.1093/emboj/19.24.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284(5411):147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 18.Wakasugi K, Schimmel P. Highly differentiated motifs responsible for two cytokine activities of a split human tRNA synthetase. J Biol Chem. 1999;274(33):23155–23159. doi: 10.1074/jbc.274.33.23155. [DOI] [PubMed] [Google Scholar]
- 19.Lee PS, et al. Uncovering of a short internal peptide activates a tRNA synthetase procytokine. J Biol Chem. 2012;287(24):20504–20508. doi: 10.1074/jbc.C112.369439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bec G, Kerjan P, Waller JP. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1 beta gamma delta complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1 delta subunit in complex formation. J Biol Chem. 1994;269(3):2086–2092. [PubMed] [Google Scholar]
- 21.Negrutskii BS, et al. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J Biol Chem. 1999;274(8):4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 22.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11(9):668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simos G, et al. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15(19):5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 24.He R, et al. Two non-redundant fragments in the N-terminal peptide of human cytosolic methionyl-tRNA synthetase were indispensable for the multi-synthetase complex incorporation and enzyme activity. Biochim Biophys Acta. 2009;1794(2):347–354. doi: 10.1016/j.bbapap.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Lee SW, et al. Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci. 2004;117(Pt 17):3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 26.Simader H, et al. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic Acids Res. 2006;34(14):3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV, et al. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain--study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3(11):2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang SS, Chang GG. Kinetic characterization of the endogenous glutathione transferase activity of octopus lens S-crystallin. J Biochem. 1996;119(6):1182–1188. doi: 10.1093/oxfordjournals.jbchem.a021366. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi S, Kidou S, Ejiri S. Detection and characterization of glutathione S-transferase activity in rice EF-1betabeta’gamma and EF-1gamma expressed in Escherichia coli. Biochem Biophys Res Commun. 2001;288(3):509–514. doi: 10.1006/bbrc.2001.5799. [DOI] [PubMed] [Google Scholar]
- 30.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc Natl Acad Sci U S A. 2002;99(12):7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rho SB, et al. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci U S A. 1999;96(8):4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quevillon S, et al. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285(1):183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 33.Kim JE, et al. An elongation factor-associating domain is inserted into human cysteinyl-tRNA synthetase by alternative splicing. Nucleic Acids Res. 2000;28(15):2866–2872. doi: 10.1093/nar/28.15.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda J, et al. Low conservation and species-specific evolution of alternative splicing in humans and mice: comparative genomics analysis using well-annotated full-length cDNAs. Nucleic Acids Res. 2008;36(20):6386–6395. doi: 10.1093/nar/gkn677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiba K. Intron positions delineate the evolutionary path of a pervasively appended peptide in five human aminoacyl-tRNA synthetases. J Mol Evol. 2002;55(6):727–733. doi: 10.1007/s00239-002-2368-3. [DOI] [PubMed] [Google Scholar]
- 36.Brenner S, Corrochano LM. Translocation events in the evolution of aminoacyl-tRNA synthetases. Proc Natl Acad Sci U S A. 1996;93(16):8485–8489. doi: 10.1073/pnas.93.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray PS, et al. Evolution of function of a fused metazoan tRNA synthetase. Mol Biol Evol. 2011;28(1):437–447. doi: 10.1093/molbev/msq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rho SB, et al. A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J Biol Chem. 1998;273(18):11267–11273. doi: 10.1074/jbc.273.18.11267. [DOI] [PubMed] [Google Scholar]
- 39.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 40.Ting SM, Bogner P, Dignam JD. Isolation of prolyl-tRNA synthetase as a free form and as a form associated with glutamyl-tRNA synthetase. J Biol Chem. 1992;267(25):17701–17709. [PubMed] [Google Scholar]
- 41.Ewalt KL, et al. Variant of human enzyme sequesters reactive intermediate. Biochemistry. 2005;44(11):4216–4221. doi: 10.1021/bi048116l. [DOI] [PubMed] [Google Scholar]
- 42.Cahuzac B, et al. A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. EMBO J. 2000;19(3):445–452. doi: 10.1093/emboj/19.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampath P, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119(2):195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 44.Arif A, et al. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol Cell. 2009;35(2):164–180. doi: 10.1016/j.molcel.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia J, et al. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29(6):679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapasi P, et al. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell. 2007;25(1):113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ilyin VA, et al. 2.9 A crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci. 2000;9(2):218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzima E, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A. 2003;100(25):14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otani A, et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A. 2002;99(1):178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sajish M, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nat Chem Biol. 2012 doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutton RB, et al. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395(6700):347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 53.Chan DC, et al. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 54.Ahn HC, kim s, lee bj. Solution structure and p43 binding of the p38 leucine zipper motif: coiled-coil interactions mediate the association between p38 and p43. FEBS Lett. 2003;542(1-3):119–124. doi: 10.1016/s0014-5793(03)00362-4. [DOI] [PubMed] [Google Scholar]
- 55.Deutscher MP, Ni RC. Purification of a low molecular weight form of rat liver arginyl-tRNA synthetase. J Biol Chem. 1982;257(11):6003–6006. [PubMed] [Google Scholar]
- 56.Vellekamp G, Sihag RK, Deutscher MP. Comparison of the complexed and free forms of rat liver arginyl-tRNA synthetase and origin of the free form. J Biol Chem. 1985;260(17):9843–9847. [PubMed] [Google Scholar]
- 57.Zheng YG, et al. Two forms of human cytoplasmic arginyl-tRNA synthetase produced from two translation initiations by a single mRNA. Biochemistry. 2006;45(4):1338–1344. doi: 10.1021/bi051675n. [DOI] [PubMed] [Google Scholar]
- 58.Shiba K, et al. Human cytoplasmic isoleucyl-tRNA synthetase: selective divergence of the anticodon-binding domain and acquisition of a new structural unit. Proc Natl Acad Sci U S A. 1994;91(16):7435–7439. doi: 10.1073/pnas.91.16.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rho SB, et al. Interaction between human tRNA synthetases involves repeated sequence elements. Proc Natl Acad Sci U S A. 1996;93(19):10128–10133. doi: 10.1073/pnas.93.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segev N, Hay N. Hijacking Leucyl-tRNA Synthetase for Amino Acid-Dependent Regulation of TORC1. Mol Cell. 2012;46(1):4–6. doi: 10.1016/j.molcel.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han JM, et al. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012 doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 62.Bonfils G, et al. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol Cell. 2012;46(1):105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Finarov I, et al. Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns. Structure. 2010;18(3):343–353. doi: 10.1016/j.str.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Dou X, Limmer S, Kreutzer R. DNA-binding of phenylalanyl-tRNA synthetase is accompanied by loop formation of the double-stranded DNA. J Mol Biol. 2001;305(3):451–458. doi: 10.1006/jmbi.2000.4312. [DOI] [PubMed] [Google Scholar]
- 65.Crepin T, et al. A hybrid structural model of the complete Brugia malayi cytoplasmic asparaginyl-tRNA synthetase. J Mol Biol. 2011;405(4):1056–1069. doi: 10.1016/j.jmb.2010.11.049. [DOI] [PubMed] [Google Scholar]
- 66.Grant TD, et al. Structural conservation of an ancient tRNA sensor in eukaryotic glutaminyl-tRNA synthetase. Nucleic Acids Res. 2012;40(8):3723–3731. doi: 10.1093/nar/gkr1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu XL, et al. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat Commun. 2012;3 doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herzog W, et al. Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ Res. 2009;104(11):1260–1266. doi: 10.1161/CIRCRESAHA.108.191718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukui H, Hanaoka R, Kawahara A. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ Res. 2009;104(11):1253–1259. doi: 10.1161/CIRCRESAHA.108.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raina M, et al. Association of a multi-synthetase complex with translating ribosomes in the archaeon Thermococcus kodakarensis. FEBS Lett. 2012;586(16):2232–2238. doi: 10.1016/j.febslet.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hausmann CD, Ibba M. Structural and functional mapping of the archaeal multi-aminoacyl-tRNA synthetase complex. FEBS Lett. 2008;582(15):2178–2182. doi: 10.1016/j.febslet.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frechin M, et al. Arc1p: anchoring, routing, coordinating. FEBS Lett. 2010;584(2):427–433. doi: 10.1016/j.febslet.2009.11.037. [DOI] [PubMed] [Google Scholar]
- 73.Karanasios E, Simos G. Building arks for tRNA: structure and function of the Arc1p family of non-catalytic tRNA-binding proteins. FEBS Lett. 2010;584(18):3842–3849. doi: 10.1016/j.febslet.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 74.Kim KJ, et al. Determination of three-dimensional structure and residues of the novel tumor suppressor AIMP3/p18 required for the interaction with ATM. J Biol Chem. 2008;283(20):14032–14040. doi: 10.1074/jbc.M800859200. [DOI] [PubMed] [Google Scholar]
- 75.Graindorge JS, et al. Role of Arc1p in the modulation of yeast glutamyl-tRNA synthetase activity. Biochemistry. 2005;44(4):1344–1352. doi: 10.1021/bi049024z. [DOI] [PubMed] [Google Scholar]
- 76.Wiltrout E, et al. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karanasios E, Boleti H, Simos G. Incorporation of the Arc1p tRNA-binding domain to the catalytic core of MetRS can functionally replace the yeast Arc1p–MetRS complex. J Mol Biol. 2008;381(3):763–771. doi: 10.1016/j.jmb.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 78.Galani K, et al. The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J. 2001;20(23):6889–6898. doi: 10.1093/emboj/20.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rieger RA, et al. Proteomic approach to identification of proteins reactive for abasic sites in DNA. Mol Cell Proteomics. 2006;5(5):858–867. doi: 10.1074/mcp.M500224-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Frechin M, Duchene AM, Becker HD. Translating organellar glutamine codons: a case by case scenario? RNA Biol. 2009;6(1):31–34. doi: 10.4161/rna.6.1.7564. [DOI] [PubMed] [Google Scholar]
- 81.Nagao A, et al. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc Natl Acad Sci U S A. 2009;106(38):16209–16214. doi: 10.1073/pnas.0907602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rinehart J, et al. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19(5):583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cook AG, et al. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461(7260):60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 84.Havrylenko S, et al. Caenorhabditis elegans evolves a new architecture for the multi-aminoacyl-tRNA synthetase complex. J Biol Chem. 2011;286(32):28476–28487. doi: 10.1074/jbc.M111.254037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Havrylenko S, et al. Methionyl-tRNA synthetase from Caenorhabditis elegans: a specific multidomain organization for convergent functional evolution. Protein Sci. 2010;19(12):2475–2484. doi: 10.1002/pro.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kerjan P, et al. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim Biophys Acta. 1994;1199(3):293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 87.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29(4):419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32(4):158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 89.Mirande M. Aminoacyl-tRNA Synthetases Complexes. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Georgetown: Eurekah; 2005. pp. 298–308. [Google Scholar]
- 90.Wolfe CL, et al. A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J Biol Chem. 2005;280(46):38870–38878. doi: 10.1074/jbc.M502759200. [DOI] [PubMed] [Google Scholar]
- 91.Lazard M, Mirande M, Waller JP. Expression of the aminoacyl-tRNA synthetase complex in cultured Chinese hamster ovary cells. Specific depression of the methionyl-tRNA synthetase component upon methionine restriction. J Biol Chem. 1987;262(9):3982–3987. [PubMed] [Google Scholar]
- 92.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30(10):569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 93.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34(5):603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 94.Ko YG, et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276(8):6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 95.Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life. 2010;62(4):296–302. doi: 10.1002/iub.324. [DOI] [PubMed] [Google Scholar]
- 96.Liu J, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30(5):846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi JW, et al. AIMP2 promotes TNFalpha-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J Cell Sci. 2009;122(Pt 15):2710–2715. doi: 10.1242/jcs.049767. [DOI] [PubMed] [Google Scholar]
- 98.Han JM, et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci U S A. 2008;105(32):11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oh YS, et al. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell. 2010;9(5):810–822. doi: 10.1111/j.1474-9726.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 100.Kwon NH, et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc Natl Acad Sci U S A. 2011;108(49):19635–19640. doi: 10.1073/pnas.1103922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaminska M, et al. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J Biol Chem. 2009;284(10):6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- 102.Han JM, Kim JY, Kim S. Molecular network and functional implications of macromolecular tRNA synthetase complex. Biochem Biophys Res Commun. 2003;303(4):985–993. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- 103.Norcum MT, Warrington JA. Structural analysis of the multienzyme aminoacyl-tRNA synthetase complex: a three-domain model based on reversible chemical crosslinking. Protein Sci. 1998;7(1):79–87. doi: 10.1002/pro.5560070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304(5):983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 105.Han JM, et al. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281(50):38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 106.Ofir-Birin Y, et al. Structural switch of lysyl-tRNA synthetases between translation and transcription. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.010. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fang P, et al. Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc Natl Acad Sci U S A. 2011;108(20):8239–8244. doi: 10.1073/pnas.1100224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang XL, et al. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc Natl Acad Sci U S A. 2002;99(24):15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang XL, et al. Gain-of-function mutational activation of human tRNA synthetase procytokine. Chem Biol. 2007;14(12):1323–1333. doi: 10.1016/j.chembiol.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kapoor M, et al. Mutational separation of aminoacylation and cytokine activities of human tyrosyl-tRNA synthetase. Chem Biol. 2009;16(5):531–539. doi: 10.1016/j.chembiol.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang XL, et al. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15(7):793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tolstrup AB, et al. Transcriptional regulation of the interferon-gamma-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J Biol Chem. 1995;270(1):397–403. doi: 10.1074/jbc.270.1.397. [DOI] [PubMed] [Google Scholar]
- 113.Shaw AC, et al. Mapping and identification of interferon gamma-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis. 1999;20(4-5):984–993. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<984::AID-ELPS984>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 114.Liu J, et al. A new gamma-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucleic Acids Res. 2004;32(2):719–727. doi: 10.1093/nar/gkh240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Q, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17(1):57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu Z, et al. Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing. Structure. 2012;20(9):1470–1477. doi: 10.1016/j.str.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 118.Xirodimas DP, et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9(3):280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jones J, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7(3):1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6(2):98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 121.Patzko A, Shy ME. Update on Charcot-Marie-Tooth disease. Curr Neurol Neurosci Rep. 2011;11(1):78–88. doi: 10.1007/s11910-010-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nangle LA, et al. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci U S A. 2007;104(27):11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Motley WW, et al. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 2011;7(12):e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xie W, et al. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2007;104(24):9976–9981. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.He W, et al. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc Natl Acad Sci U S A. 2011;108(30):12307–12312. doi: 10.1073/pnas.1104293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Froelich CA, First EA. Dominant Intermediate Charcot-Marie-Tooth disorder is not due to a catalytic defect in tyrosyl-tRNA synthetase. Biochemistry. 2011;50(33):7132–7145. doi: 10.1021/bi200989h. [DOI] [PubMed] [Google Scholar]
- 127.Storkebaum E, et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc Natl Acad Sci U S A. 2009;106(28):11782–11787. doi: 10.1073/pnas.0905339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jordanova A, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38(2):197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 129.Antonellis A, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72(5):1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guo M, et al. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: Implications for multisynthetase complex formation. Proc Natl Acad Sci U S A. 2008;105(7):2331–2336. doi: 10.1073/pnas.0712072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McLaughlin HM, et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87(4):560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vo MN, Yang XL, Schimmel P. Dissociating quaternary structure regulates cell-signaling functions of a secreted human tRNA synthetase. J Biol Chem. 2011;286(13):11563–11568. doi: 10.1074/jbc.C110.213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kleiman L, Jones CP, Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010;584(2):359–365. doi: 10.1016/j.febslet.2009.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kovaleski BJ, et al. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J Biol Chem. 2006;281(28):19449–19456. doi: 10.1074/jbc.M601189200. [DOI] [PubMed] [Google Scholar]
- 135.Guo M, et al. Packaging HIV Virion Components through Dynamic Equilibria of a Human tRNA Synthetase. J Phys Chem B. 2010;114(49):16273–16279. doi: 10.1021/jp1082517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Harris JK, et al. The genetic core of the universal ancestor. Genome Res. 2003;13(3):407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]