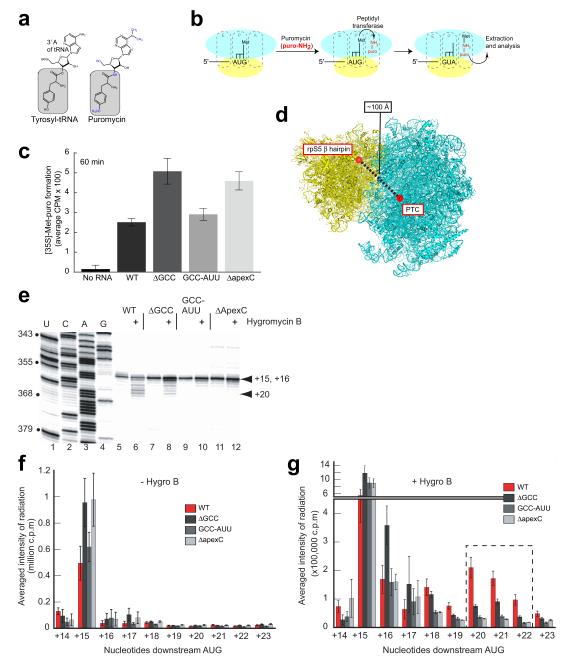
Figure 6. Puromycin and toeprinting assays with antiobiotic.
(a) Structures of tyrosyl-tRNA (left) and the puromycin (right), differences in grey and blue. (b) Cartoon of the puromycin assay (40S subunit yellow, 60S blue) moving from left to right: IRES-40S formation, then Met-tRNAi and puromycin (puro-NH2) binding in the 60S subunit P and A sites, respectively. Peptidyl transferase results in methionine bound to puromycin via a noncanonical amide linkage (met-puro), then extraction. (c) Quantitated and background-corrected graph of met-puro formation after 60 minutes on wild-type (WT) and mutant IRES RNAs. Error bars: one s.e.m of three independent duplicate experiments. (d) Yeast 80S ribosome crystal structure 56 (40S subunit yellow, 60S cyan) with approximate locations and distance between the IRES domain IIb (dIIb)-rpS5 interaction and the peptidyl transferase center (PTC) shown. (e) Relevant part of the toeprint gel with the dideoxy sequencing reactions in lanes 1-4, free IRES-80S complexes in rabbit reticulocyte lysate (RRL) without any antibiotic in lanes 5, 7, 9 and 11 as well as initiating and elongating IRES-80S complexes formed in RRL with hygromycin B in lanes 6, 8, 10 and 12. Black arrowhead represents initiating complexes (+15, +16) and blue arrowhead represents elongating complexes (+20) on the right, nucleotide numbers are bulleted, on the left. (f) Graph of quantitated, normalized, and background-corrected toeprints without antibiotic from panel e (WT IRES red, dIIb mutants grey). (g) Same as in panel f except graph represents toeprints with antibiotic. Error bars represent one s.d. of three independent experiments.