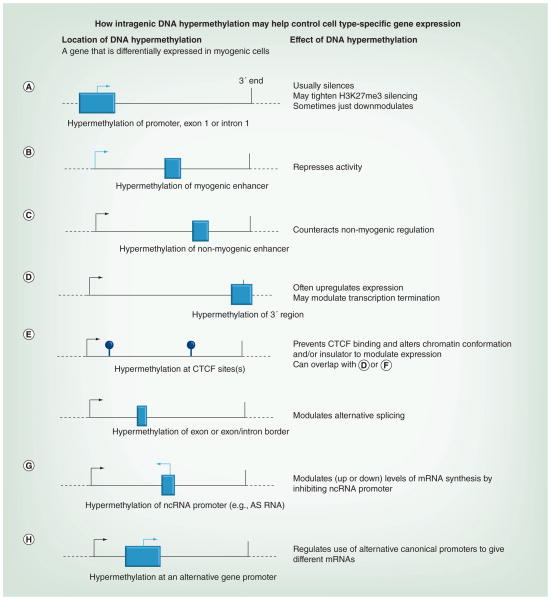
Figure 3. Model for the differentiation-associated regulation of gene expression by DNA hypermethylation.
Some known or proposed types of DNA methylation-dependent control of gene expression using the example of myogenesis-associated DNA hypermethylation are shown. The model depicts just cis-acting regulation from within the gene or from its canonical promoter. DNA methylation changes may be stabilizing and/or initiating the depicted changes in transcription. Boxes show hypermethylated differentially methylated regions. Light arrows indicate inhibited transcription start sites and black arrows indicate active transcription start sites that were not silenced by DNA methylation. (A) Hypermethylation of the promoter region, exon 1 or intron 1. (B) Hypermethylation of a myogenic enhancer in nonmyogenic cells. (C) Hypermethylation of a non-myogenic enhancer in myogenic cells. (D) Hypermethylation of the 3′ region. (E) Hypermethylation at CTCF sites (lollipops) somewhere in the gene. (F) Hypermethylation in an exon or at an exon/ intron border. (G) Hypermethylation of an intragenic ncRNA promoter. (H) Hypermethylation of an alternative promoter for a gene.
AS: Antisense.