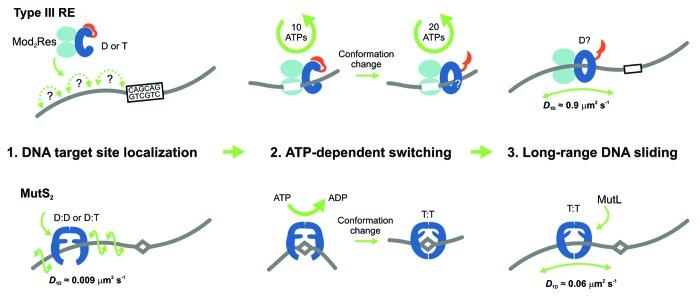
Figure 1. Models for the ATP-dependent initiation of DNA sliding by type III restriction endonucleases5 (upper) and the MutS enzymes of mismatch repair6-8 (lower). The type III restriction enzyme EcoP15I comprises 2 Mod subunits (methyltransferase) and 1 Res subunit (helicase in blue, endonuclease in red). MutS enzymes are either homo- or heterodimers. (D = ADP, T = ATP). (1) EcoP15I binds to its target site (5′-CAGCAG-3′), predominantly by a 3D hopping or jumping search pathway, whereas MutS enzymes search for DNA damage by 1D DNA sliding correlated with the DNA helix. (2) Both enzyme systems form a stable complex with their respective target site. The role of ATP is to trigger site release and cause a conformational switch into the activated sliding state necessary for downstream signaling and recruitment of protein co-factors. While EcoP15I requires hydrolysis of ~30 ATPs for this process, MutS proteins require an exchange of 1 or 2 bound ADPs for ATP. While MutS structures indicate how sliding clamps could form, structural information on the type III enzymes is currently lacking. (3) Both enzyme systems enter into a long-lived 1D sliding state. Motion of both classes of enzyme is uncorrelated with respect to the DNA helix, which probably reflects that the downstream interactions are via protein–protein contacts rather than protein–DNA contacts. The MutS diffusion is faster in this step than during the initial mismatch search.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.