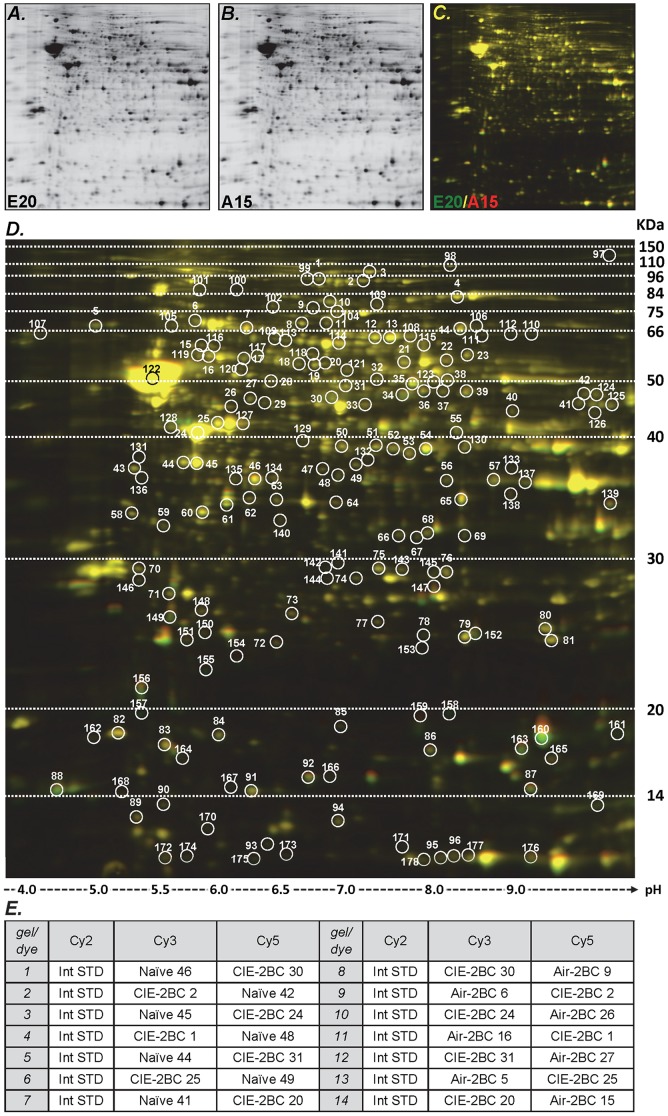
Figure 2. Example of 2D-DIGE gel. Gel images were scanned immediately following the SDS-PAGE.
Each scan revealed one of the CyDye signals (Cy2, Cy3 and Cy5). Cy2 was used to normalize the signals from Cy3 and Cy5 channels. Overlay images were generated to compare different samples. The images shown were obtained from gel#14, CTX samples. A: proteome from sample E20, labeled with Cy3; B: proteome from sample A15, labeled with Cy5; C: overlay image generated from gel#14. In this case, a green spot represents an up-regulated protein in the CIE-2BC sample E20, compared to the Air-2BC sample A15. D: Overlay image and spot selection. Single and overlay images were generated, and a comparative analysis of all spots was performed using DeCyder “in-gel” or “cross-gel” analysis software. Spots 1-96 were selected based on 1.25-fold and p<0.1 for protein ratio cut-off, allowing for the appearance of the spots in 27 out of 28 gels (81 out of 84 total images). Spots 97-178 were added by lowering the cut-off to 1.15-fold for the significance threshold, without restricting the spots based on p-values; also, the stringency was lowered so that the spots appeared in at least 23 out of 28 gels (69 out of 84 appearances). E: Experimental design for 2D-DIGE gels. Two samples and the internal standard (Int STD) were loaded in each gel. Fourteen gels were run with CTX samples and an additional fourteen gels were run with MB samples. The randomized sample order and the dye-swap design avoided experimental biases.