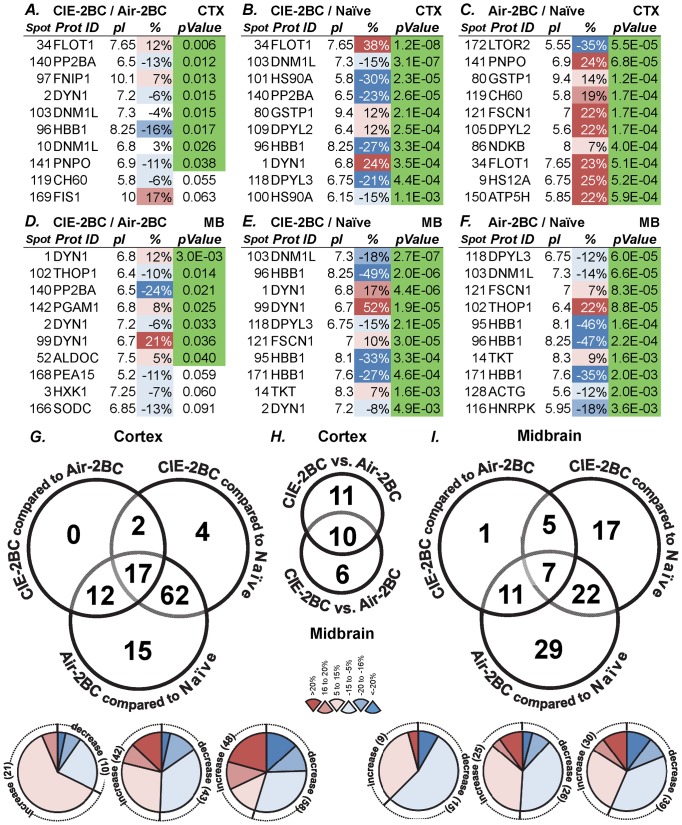
Figure 3. Protein expression changes induced by CIE paradigm.
A-F, top 10 significant differentially expressed proteins for each comparison and brain region analyzed. A-C, cortex; D-F, midbrain. A, D: CIE-2BC vs. Air-2BC; B, E: CIE-2BC vs. Naïve; C, F: Air-2BC vs. Naïve. Different isoforms are listed for some proteins. Prot ID, protein ID; pI, isoelectric point. Refer to Methods for p-values and to Table S1 for the complete list of proteins. G-H, number of spots with differentially expressed proteins across specific group comparisons for CTX (G), MB (I), and between them (H). Number and direction of protein expression changes between CIE-2BC versus Air-2BC, CIE-2BC versus Naïve, and Air-2BC versus Naïve groups. The Venn diagrams indicate the number of shared and unique protein spots among comparisons. The pie charts show the amount of protein that increases and decreases within three ranges: 5-15%, 16-20%, >20% (absolute values); the numbers in brackets indicate the number of protein spots in that category. A total of 178 spots with stringency of 69/84 gels were considered. Only differences greater than 1.05 fold with p<0.2 are listed (Student's t-test). Most of the corresponding proteins have been identified.