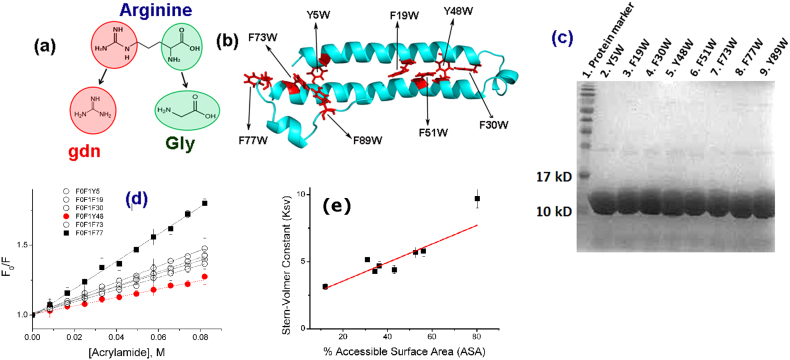
Figure 1.
(a) The molecular structure of arginine: The backbone resembles glycine, which is a known protecting osmolyte. The side chain of arginine is identical to gdn.HCl, which is a common denaturant. (b) The predicted structure of KMP-11: The positions of the inserted tryptophan residues are shown using black arrows. The predicted structure was obtained using a combination of threading, ab-initio modeling and atomic-level structure refinement methods of I-TASSER28,29 from Zhanglab server. The modeled structure was equilibrated in aqueous environment using a molecular dynamic simulation run of 100 nsec (see materials and methods, Supporting Information); (c) SDS page electrophores of the purified mutants (d) Acrylamide quenching experiments of the tryptophan mutants of KMP-11: The change in F0/F with acrylamide concentration is the minimum for the Y48W mutant (red line with filled circles). The change in F0/F with the acrylamide concentration is the maximum for the F77W mutant (black line with filled squares). The change in F0/F with acrylamide concentrations for other mutants lies between these two extremes (black lines with hollow symbols); (e) The linear correlation observed between the experimentally determined KSV (M−1) values and the calculated solvent accessible surface area (SASA) (%). Three independent measurements were used to calculate the error bars for Ksv.