Figure 1.
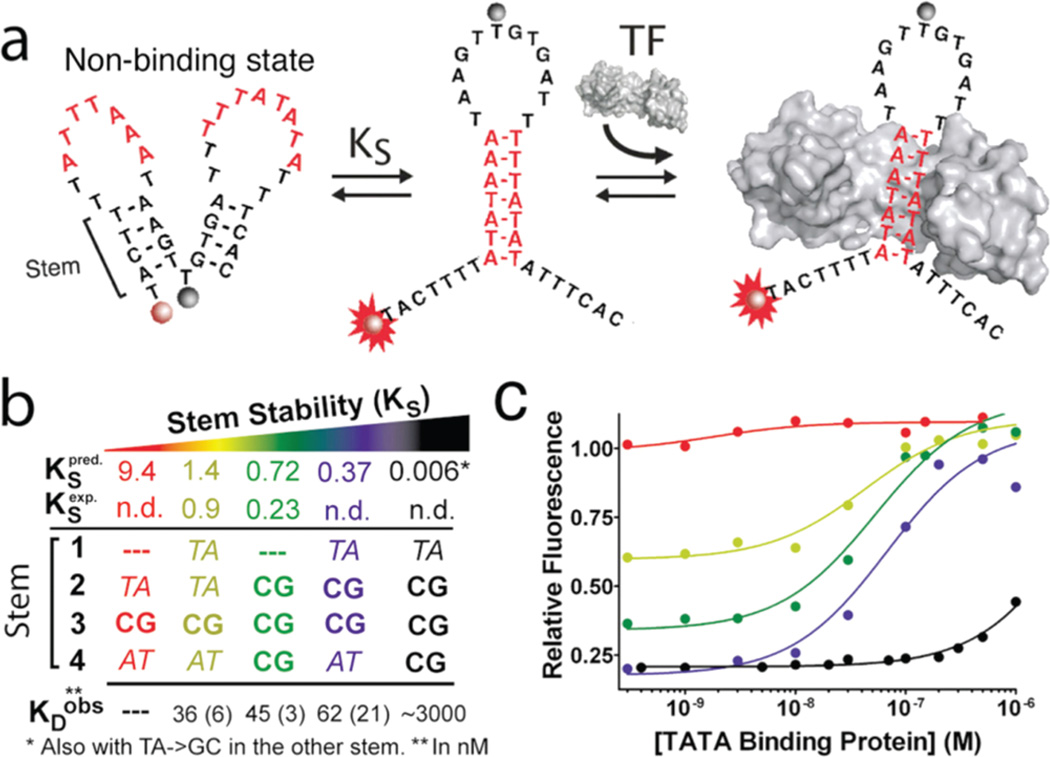
Transcription factor (TF) beacons for the quantitative detection of DNA binding activity. (a) DNA sequences containing the recognition site for a specific DNA binding protein (here shown, red stem, for TATA binding protein (TBP)) are engineered into switches by stabilizing an alternative “non-binding” conformation or state (left). Binding of the protein thus shifts the switch’s conformational equilibrium toward the binding-competent state, which, in turn, is linked to an increase in fluorescence. (b and c) Optimal detection limits are achieved at intermediate values of the switching equilibrium constant (KS) as this produces a switch that, in the absence of target, is predominantly in its dark “non-binding” state without overstabilizing it, which would reduce the beacon’s affinity (Supporting Figure 1).17