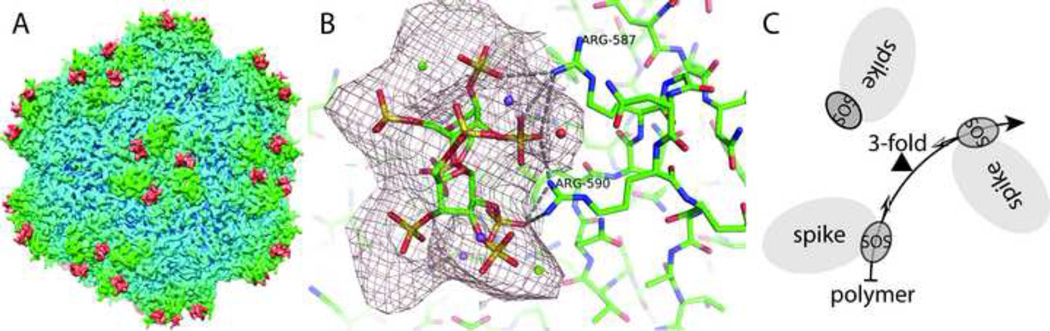
Figure 1.
3D cryo-EM reconstruction of AAV-DJ complexed with SOS at 4.8 Å resolution. A: Peaks (segmented in red) are seen at the SOS binding sites in the map of the complex on the side of each spike that surrounds each viral 3-fold axis. B: The Δρ difference map (complex – native), contoured at 6σ shows, the SOS sulfate locations clearly, even at this resolution. Three of the sulfates appear to form salt bridge interactions with Arg587 and Arg590 from one of the exterior loops of the virus structure. C: Schematic of the central part of panel A showing the binding sites on three adjacent spikes related by icosahedral symmetry. As indicated by the two-tone arrows emanating from each SOS and pointing towards the 3-fold axis (triangle), a disaccharide could be bound at each site in the same orientation with respect to the local virus environment. The 8 Å resolution structure of the AAV-2 heparin complex (O'Donnell et al., 2009) shows diffuse density bridging between adjacent sites, consistent with a single polysaccharide passing through both sites. In such a case, the direction of the polysaccharide backbone in one site, relative to the local protein environment, would be the opposite of the direction at the adjacent site. This suggests that a virus that had evolved for polyvalent binding would have local binding sites tolerant of two GAG orientations.