FIGURE 6.
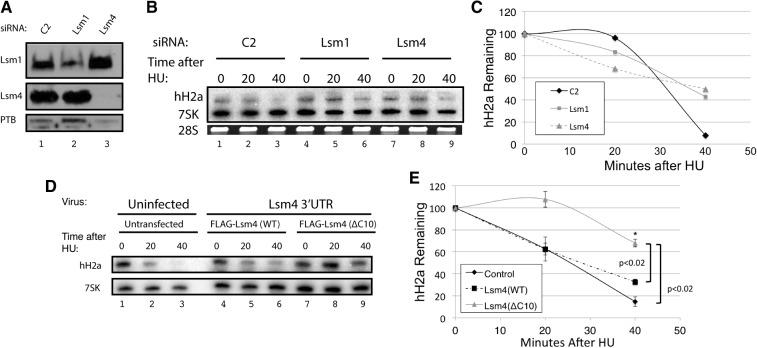
The interaction of Lsm4 with SLBP and 3′hExo is necessary for degradation of histone mRNA after inhibiting DNA replication. (A) HeLa cells were treated with a control siRNA (lane 1), an siRNA targeting Lsm1 (lane 2), and an siRNA targeting Lsm4 (lane 3), as previously described (Mullen and Marzluff 2008). Total cell lysates were prepared and analyzed by Western blotting for Lsm1 (top), Lsm4 (middle), and polypyrimidine track binding protein (PTB, bottom). (B,C) The cells in panel A (exponentially growing) were treated with 5 mM HU, and RNA was harvested before HU treatment and 20 and 40 min after HU treatment (HU). Total cell RNA was purified, and 1 μg of whole cell RNA was resolved on 8 M urea–6% polyacrylamide gels, and histone H2a mRNA and 7SK RNA were detected by Northern blotting and visualized with a PhosphorImager. The RNA samples were also analyzed by gel electrophoresis and stained with ethidium bromide for 28S rRNA (bottom). (C) The PhosphorImager images were analyzed by ImageQuant. (D,E) Exogenous Flag-Lsm4 with a wild-type C-terminal tail or Flag-Lsm4(ΔC10) was stably expressed in HeLa cells. Endogenous Lsm4 was knocked down by an shRNA targeting the 3′ UTR of endogenous Lsm4 (Fig. 5C). Histone mRNA degradation was initiated by treating exponentially growing cells with 5 mM HU, and the total cell RNA was analyzed as in panel B. The results of three independent experiments were quantified in panel E; statistical significance was determined by a Student's t-test.