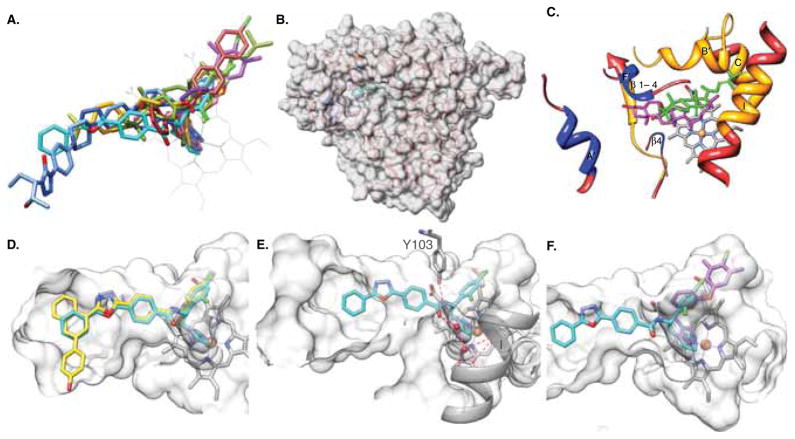
Figure 2. T. cruzi CYP51 structure as a template for rational drug design.
The orientation of the protein in all panels is similar (~ distal view of a P450 molecule). A. CYP51-structure-based inhibitory pharmacophore including imidazoles VNI (cyan, PDB ID [3gw9]), VNF (brick, [3skw]), and NEE (purple, [4h6o]); triazoles posaconazole (blue, [3k1o]), and fluconazole (pink, [3lfd]); pyridine derivative UDO (goldenrod, [3zg2]) and substrate analog MCP (olive, [3p99]). B. Overall semitransparent surface representation of the CYP51 molecule, the enlarged view of the secondary structural elements forming the substrate access channel and the binding cavity is seen in c. C. The three secondary structural elements forming the entrance into the substrate binding channel are colored in blue. The CYP51 binding cavity (orange) protrudes deeper inside the enzyme molecule. T. cruzi CYP51 substrate eburicol (olive) is shown in comparison with cholesterol (magenta) in the superimposed CYP11A1 [3mzs]. The 14α atom of eburicol is positioned directly above the heme iron, while in CYP11A1, the P450 which is functionally closest to CYP51 and also catalyzes a three-step reaction cleaving the side chain of cholesterol at C22; it is the cholesterol C22 atom that is positioned above the hem. D. A hypothetical three-ring structure (yellow) that may fit into the area around the entrance into the CYP51 substrate access channel. E. VNI in the CYP51 substrate binding cavity; the hydrogen bond network is shown as red dashed lines. F. VNI and NEE in the superimposed CYP51 co-structures showing that the VNI β-phenyl ring arm can be elongated. The binding cavity in d, e, and f, d, is shown in semitransparent surface representation.