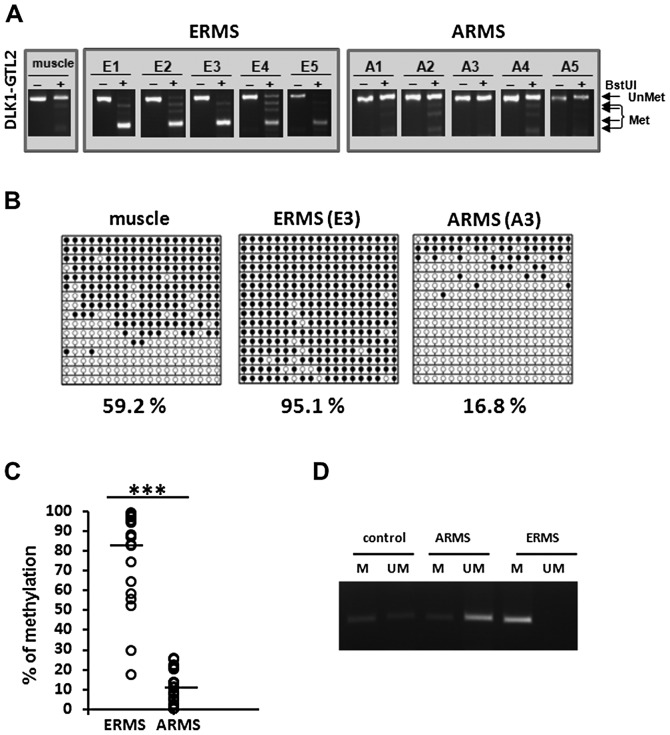
Figure 1.
The methylation pattern of the IG-DMR at the DLK1-GTL2 locus differs between ARMS and ERMS patient samples. (A) The COBRA assay was used to evaluate the methylation status of the IG-DMR at the DLK1-GTL2 locus by employing the restriction enzyme BstUI. Methylated DNA (Met) is cleaved because methylcytosine is not deaminated to uracil by bisulfite treatment and thus the BstUI recognition site (CGCG) is maintained. By contrast, unmethylated DNA (UnMet) is not cleaved because the cytosine is deaminated to uracil and thus the sequence of the recognition site is changed. (B) An example of results from bisulfite modification of DNA followed by sequencing to evaluate DNA methylation of the IG-DMR at the DLK1-GTL2 locus in human skeletal muscle samples and two RMS samples (ERMS and ARMS). Methylated and unmethylated CpG sites are shown as filled and open circles, respectively. Each horizontal line represents one subclone. The numbers under the bisulfite sequencing profiles indicate the percentage of methylated CpG sites. (C) Densitometric analysis of the methylation of the IG-DMR at the DLK1-GTL2 locus based on the COBRA assay results for 22 ERMS and 23 ARMS samples. (D) Methylation-specific PCR analysis of the IG-DMR at the DLK1-GTL2 locus in normal tissue (muscle) and two RMS samples (ERMS and ARMS). UM, unmethylated alleles; M, methylated alleles.