Table 4.
Indicating average weights of ten top important features computed by MCRLR in the jackknife procedure using all of features for prediction of five RNA-binding domain subclasses. Positive values show preference of the features in related subclasses and negative values show avoidance of the features in related subclasses.
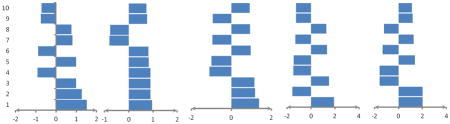
| |||||
|---|---|---|---|---|---|
| No. | Feature/Average Value
|
||||
| 7s RBD subclass | Double -stranded RBD subclass | mRNA RBD subclass | rRNA RBD subclass | tRNA RBD subclass | |
| 1 | Number of Met in MP/1.517 | Number of basic AAs in MP/0.967 | Molecular weight/1.373 | Number of His in MP/1.913 | Number of Arg in IR/2.004 |
| 2 | Number of Met in RC/1.274 | Number of Glu in MP/0.900 | Number of Ser in BR/1.176 | Number of Arg in IR/-1.555 | Number of His in sheet/2.003 |
| 3 | Number of Cys in sheet/0.998 | Dipole/0.900 | Number of Cys in MP/1.142 | Number of Ile in MP/1.499 | Number of Pro in MP/-1.583 |
| 4 | Number of His in seq./-0.920 | Number of Glu in IR/0.891 | Number of Ser in IR/-1.094 | Number of Leu in helix/-1.481 | Number of Ile in MP/-1.576 |
| 5 | Number of Met in ER/0.990 | Number of Tyr in MP/0.831 | Isoelectric point/-1.007 | Number of Met in RC/-1.458 | Number of Asp in BR/1.371 |
| 6 | Number of Glu in MP/-0.886 | Number of Ala in BR/0.811 | Number of Cys in RC/0.969 | Number of Ser in RC/1.330 | Number of Ile in IR/-1.335 |
| 7 | Number of Tyr in helix/0.811 | Number of Gln in RC/-0.799 | Number of charged AAs in seq./-0.949 | Number of Tyr in MP/-1.304 | Number of Met in sheet/1.255 |
| 8 | Number of Glu in IR/0.759 | Number of Arg in MP/-0.779 | Number of Asp in ER/0.934 | Number of Val in helix/1.280 | Number of Phe in ER/-1.226 |
| 9 | Frequency of antiparallel HB/-0.753 | Number of Lys in MP/0.752 | Number of Arg in ER/-0.931 | Number of His in ER/-1.269 | Number of Phe in IR/1.166 |
| 10 | Second patch size/-0.729 | Number of Asp in MP/0.728 | Number of Asp in IR/0.920 | Number of Cys in RC/-1.258 | Number of Gln in IR/1.114 |
MP: main patch, RC: randomcoil, ER: exposed regions, IR: intermediate regions, HB: hydrogen bond, AAs: amino acids, BR: buried regions, Seq.: Sequence.
