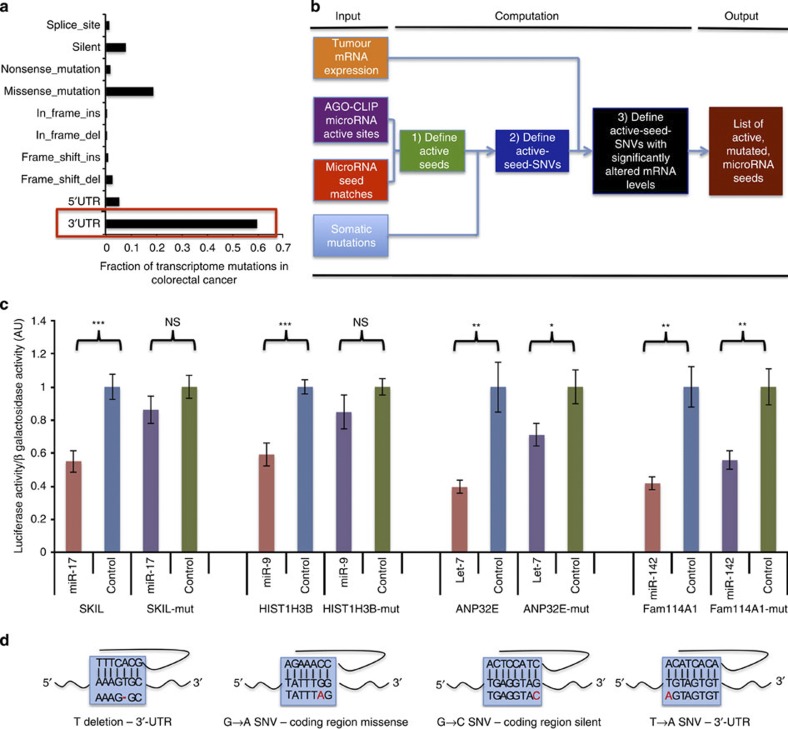
Figure 6. The miSNP algorithm defines mutations in microRNA seeds.
(a) 3′-UTR mutations make a disproportionate number of all mutations effecting mRNAs. (b) Work flow of miSNP algorithm, which integrates TCGA mutation and mRNA expression analysis and AGO-CLIP seed nominations to determine mutations in active microRNA seeds. (c) Validation of selected AGO-CLIP-defined seed SNVs demonstrates the ability for endogenous tumour mutations to ablate microRNA regulation in a predictable, seed-dependent manner. (d) Mutations reproduced in each 3′UTR construct matching endogenous somatic mutations found in the TCGA pan-cancer data as they relate to the cognate microRNA seed. *P<0.05, **P<0.005, NS, not significant; values measured with Student’s t-test. Assays were performed twice at 5 nM mimic and twice at 10 nM in quadruplicate. Results were combined for final analysis (n=16). Error bars are s.e.m.