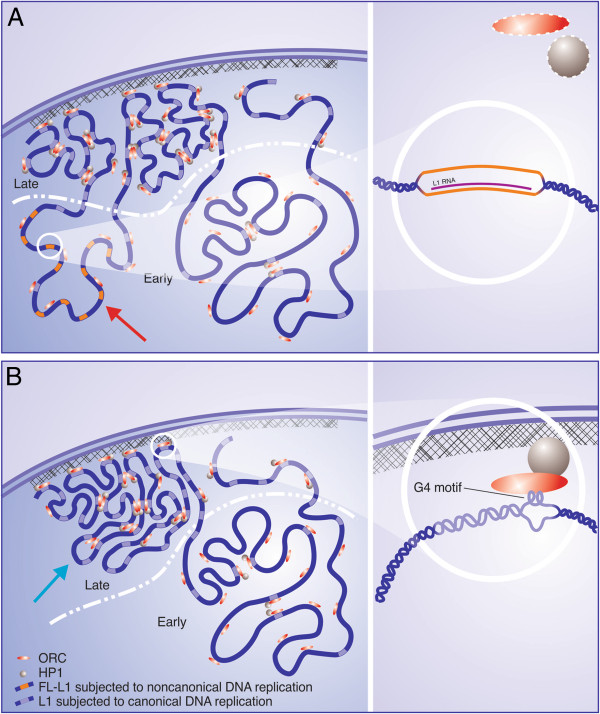
Figure 2.
A hypothetical model of two modes of replication of full-length L1s as an epigenetic switch. A: Undifferentiated (early embryonic and cancer) cells. A subset of small L1-rich domains of the genome (EtoL domains as per Hiratani and co-authors [45]) replicates DNA in early S (red arrow). Transcriptional competence is imposed on these domains during early S phase of DNA replication. These L1-rich EtoL domains do not tether to the nuclear lamina and have a loose conformation of chromatin loops. Genes residing in these domains are linked to undifferentiated states of a cell. Noncanonical replication of FL-L1s residing in these domains might prevent them from binding to the nuclear matrix and from recruiting the ORCs (panel A, right) and, therefore, from being silenced through the ORC/HP1-mediated pathway of heterochromatin assembly. B: Differentiated and differentiating cells. Global programmed downregulation of FL-L1s upon differentiation of pluripotent embryonic cells results in switching “off” the noncanonical mechanism of L1 DNA replication. In the absence of L1 RNA paired with a complementary “parental” L1 DNA for noncanonical L1 DNA replication, L1s might attach to the nuclear matrix and form intrastrand G4 structures. The majority of replication origins are associated with G4s [80]. The L1-bound ORC might bind HP1 in a distinct chromatin environment and, thereby, play a crucial role in the establishment of a silent state on the L1-rich/gene-rich EtoL domains (panel B, right). These domains switch to their default state characterized by late replication, dense conformation, and tethering to the nuclear periphery [50] (blue arrow). Basically, the same switch from noncanonical to canonical replication of L1s residing within EtoL domains might occur upon differentiation of poorly differentiated cancer cells caused by the knockdown of the expression of L1s.