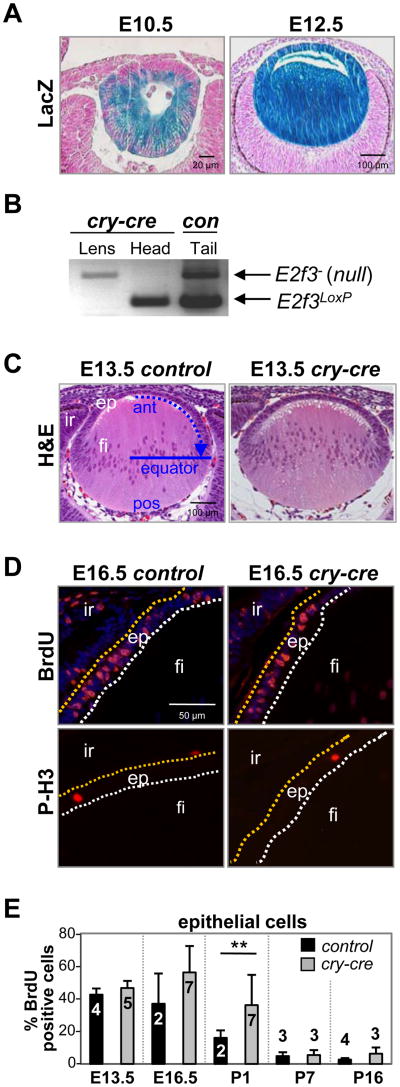
Fig. 1. Conditional deletion of E2f3 in cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses does not block epithelial cell proliferation.
(A) Cre recombinase activity in cry-cre;ROSA26Sortm1Sor mice is detectable as a blue stain (LacZ expression) in embryonic lens epithelial and fiber cells. (B) PCR genotyping of E2f3 on lens or head tissue collected by laser capture microdissection. Control tail DNA was collected from a weanling E2f3LoxP/- mouse. (C) E13.5 lenses of E2f1-/-;E2f2-/-;E2f3LoxP/LoxP(control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) mice were stained by H&E to examine nuclear distribution and lens architecture. Blue arrow indicates direction of epithelial migration toward equatorial line; ant, anterior; pos, posterior; ep, epithelium; fi, fiber cells; ir, iris. (D) Immunodetection of BrdU incorporation and phosphorylated histone H3 show normal patterns of DNA synthesis and proliferation in E16.5 control and cry-cre lenses. Epithelial cells positive for BrdU or phosphohistone H3 stain red. Nuclei are stained by DAPI in blue (top quadrants). (E) The percentage of BrdU-positive epithelial cells in lenses at the indicated ages. A minimum of three sections near the central plane of cry-cre lenses were analyzed at E13.5 (n=5), E16.5 (n=7), P1 (n=7), P7 (n=3), and P16 (n=3) and of control lenses at E13.5 (n=4), E16.5 (n=2), P1 (n=2), P7 (n=3), and P16 (n=4). Error bars represent standard deviation and significance of unpaired T-test is indicated by ** p < 0.01.