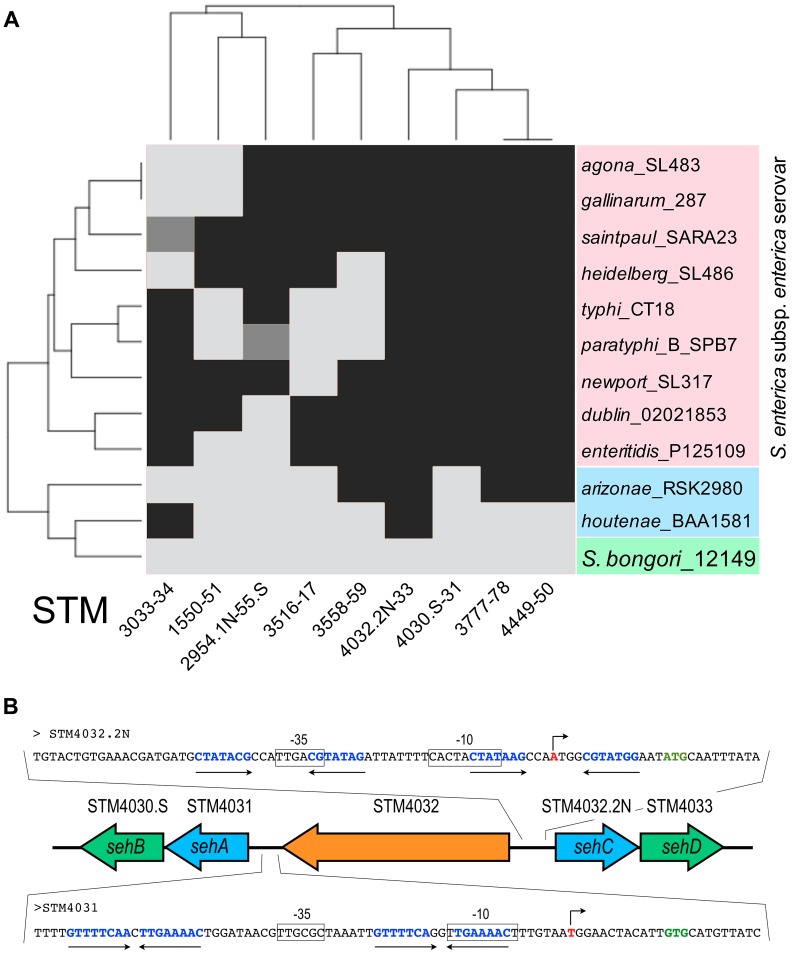
Figure 1. Conservation of TA modules across the genus Salmonella.
(A) The toxins of S. Typhimurium TA modules were subjected to BLASTP against various Salmonella strains. Serovars of S. enterica subsp. enterica are coloured in red. Arizonae and houtenae subspecies of S. enterica are coloured in blue. The non-pathogenic species bongori is coloured in green. All sequences were taken of NCBI and ColiBASE databases. Gene products were clustered according to their presence (black) or absence (light grey). Incomplete modules are indicated in dark grey. (B) Genetic organization of the STM4031-STM4030.S and STM4032.2N-STM4033 modules in S. Typhimurium chromosome. Toxin genes are coloured in blue and antitoxin genes in green. A single gene, STM4032 (in orange), separates the two TA modules. The promoter regions of the TA operons are shown as a blow-up. Putative transcriptional start sites are shown as broken arrows. Start sites of the first gene in the TA operons are shown in green bold letters. Putative −10 and −35 sequences are boxed. Palindromic sequences that are putative antitoxin binding sites are shown as opposing arrows.