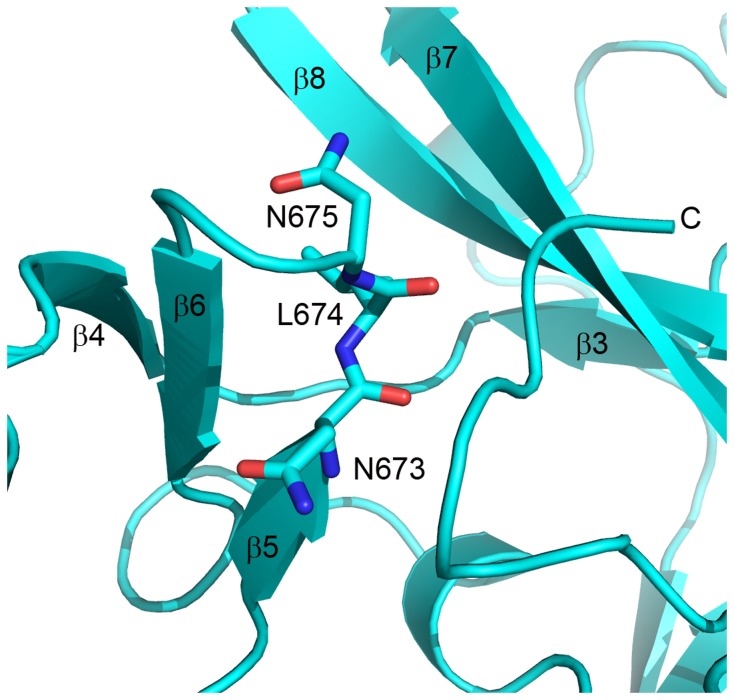
Figure 5. Mutations in the β-5/β-6 loop of EZH2-SET are contiguous with the putative substrate binding cleft.
The crystal structure of the EZH2-SET domain is represented as a ribbon model (cyan). Side chains are represented as sticks colored by atom (carbon, cyan; oxygen, red; nitrogen, blue). Secondary structure elements are labeled. N673, L674, and N675 all interact directly with the C-terminal tail which occupies the substrate binding groove. Mutation of these residues could potentially affect substrate binding in the active state as well as the transition from the inactive to active state. An N673S mutation has been identified in CMML [32]. L674V mutations have been found in both MDS [28] and AML [29]. An N675K mutation was discovered in RCMD [28].