Figure 5. In silico analysis predicted differential binding patterns for rs640249 and rs641563 polymorphisms. Gel shift experiment has confirmed this differential binding for rs640249 polymorphism.
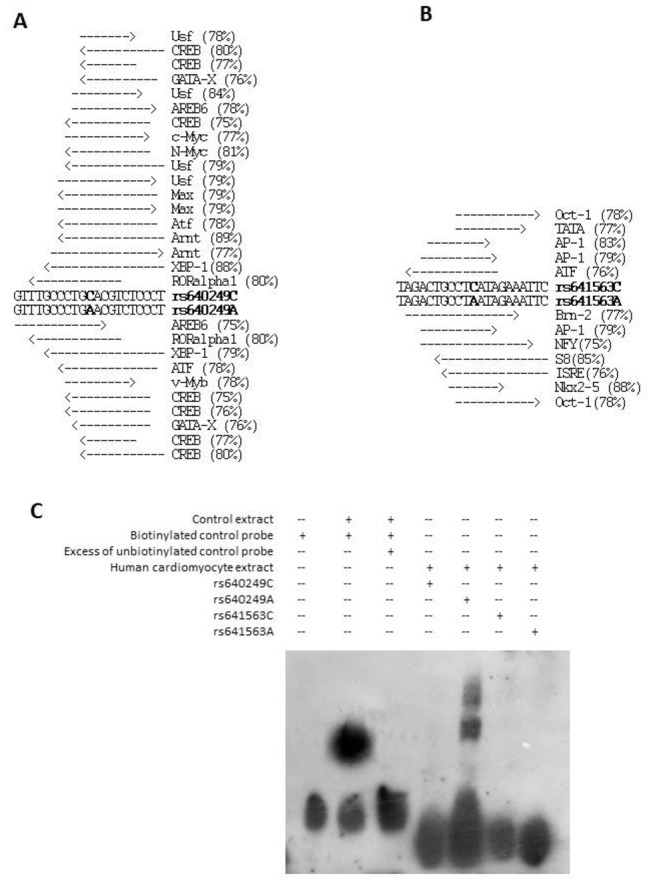
The probability of these polymorphisms creating or altering DNA–protein interaction was determined by in silico analysis (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi). An 75% threshold score (similarity matrix) was used. For each polymorphism (A, rs1800925; B, rs641563) putative bindings are described. The similarity matrix score is indicated in brackets. C: Electrophoretic mobility shift assays were performed in vitro as described in Materials and Methods. A differential binding pattern was detected for the rs640249A allele.
