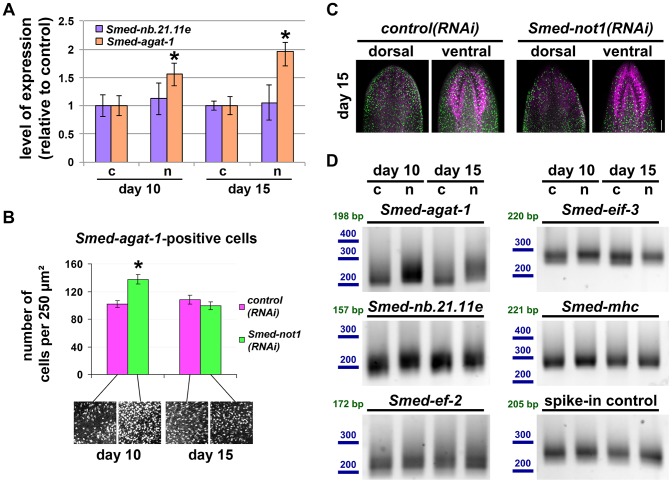
Figure 5. Smed-not1(RNAi) animals have increasing numbers of Smed-agat-1 transcripts with increased frequency of long poly(A) tails but decreasing numbers of Smed-agat-1-positive cells.
(A) Quantification of the level of expression by qRT-PCR of the progeny markers Smed-nb.21.11e and Smed-agat-1 in Smed-not1(RNAi) animals 10 and 15 days after RNAi, normalized expression and relative to respective control(RNAi) samples. Error bars represent standard deviation, asterisks represent statistical significance. Smed-agat-1 transcripts accumulate progressively after 10 and 15 days of RNAi. (B) Quantification of the number of Smed-agat-1-positive cells in control(RNAi) and Smed-not1(RNAi) worms 10 and 15 days after RNAi. Animals (N = 8 per time point and treatment) were stained by FWMISH of Smed-agat-1 and analyzed by confocal microscopy. Multiple squares corresponding to 250 µm2 in both the dorsal and ventral parts of the animals were selected for counting along the length of the animals. 130 squares were counted, 4 representative squares are shown. Error bars represent standard error of the mean, asterisk represents statistical significance. A significant accumulation of Smed-agat-1-positive cells is detected in Smed-not1(RNAi) animals 10 days after RNAi but a decrease is observed at 15 days (C) Confocal Z-projection of FWMISHs of Smed-agat1 in control(RNAi) (top panels) and Smed-not1(RNAi) (bottom panels) animals corresponding to anterior dorsal (left) and anterior ventral (right) regions of representative worms. Smed-agat-1-positive cells are shown in green, nuclei counterstaining in magenta. Scale bar: 100 µm. (D) PAT assays reflecting the distribution of mRNA poly(A) tail lengths for Smed-agat-1 and Smed-nb.21.11e, the housekeeping mRNAs Smed-ef-2 and Smed-eif-3, the tissue specific mRNA Smed-mhc and a spiked-in control mRNAs in control(RNAi) (c) and Smed-not1(RNAi) (n) animals 10 and 15 days after RNAi. Size markers used are represented in blue, the theoretical length of the amplicon corresponding to the deadenylated mRNA species given the primers used in each assay is given in green. Products above this length originate from polyadenylated mRNA molecules. Marked differences in poly(A) tail length distribution are detected for Smed-agat-1, showing an increased frequency of long poly(A) tails after Smed-not1 knock down. Slight differences are observed for Smed-nb.21.11e and Smed-eif-3.