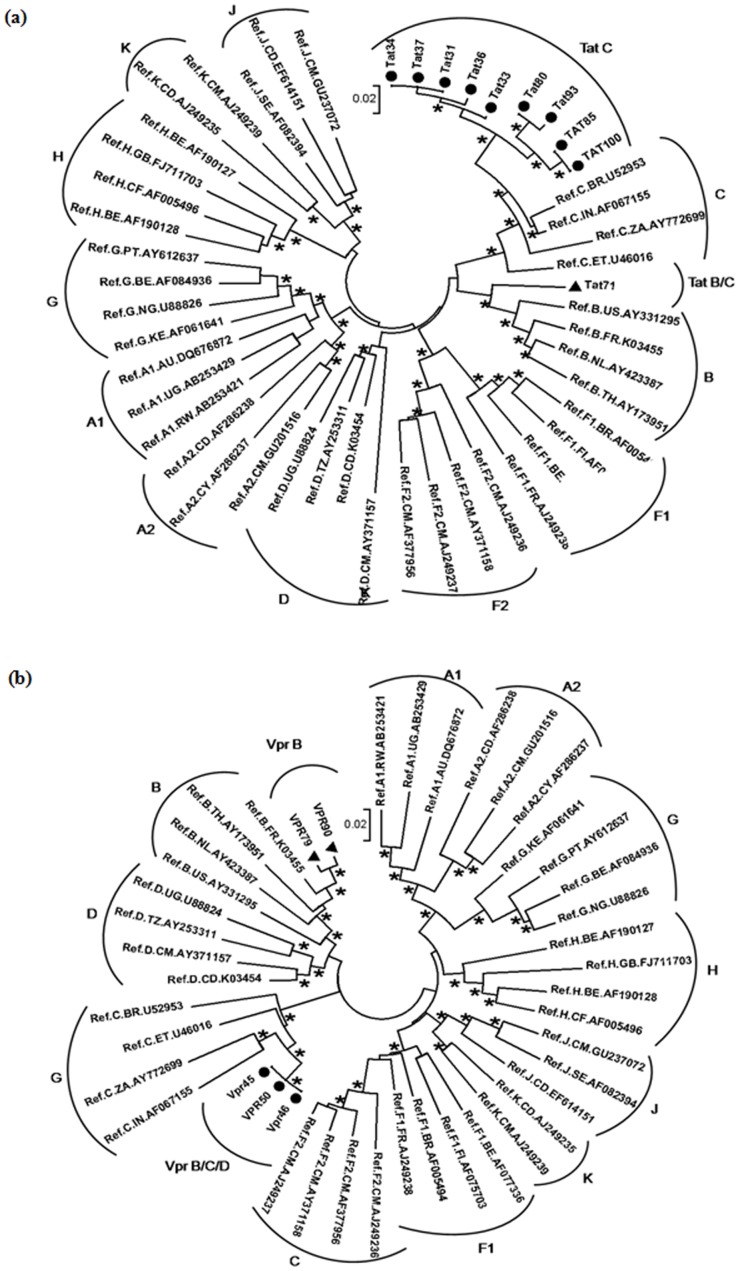
Figure 2. HIV-1 subtyping based on global subtype references.
Phylogenetic analysis was performed for Tat exon-1 and Vpr variants with M (A to K including A1, A2, F1, and F2) group. Each reference sequence was labelled with subtype, followed by country of isolation and accession number. The bootstrap probability (>65%, 1,000 replicates) was indicated with asterisk (*) at the corresponding nodes of the tree and the scale bar represents the evolutionary distance of 0.02 nucleotides per position in the sequence. (a) It represents the phylogenetic tree of Tat exon-1 variants with M group. The filled circles represent C variants and the filled triangles represent recombinants. (b) It represents the phylogenetic tree of Vpr variants with M group. The filled triangles represent B variants and the filled circles represent recombinants.