FIGURE 2.
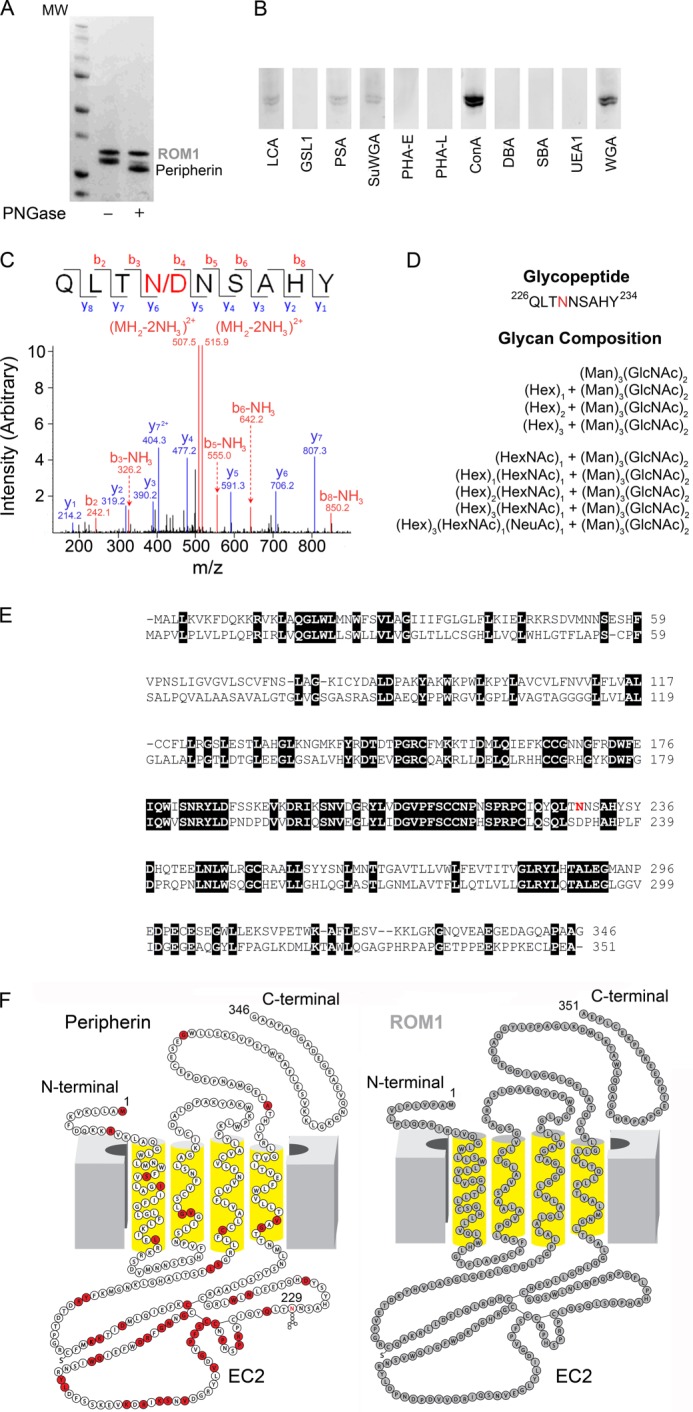
Identification of glycosylation species by mass spectrometry and comparison of peripherin and ROM1 sequences and two-dimensional structures. A, SDS-PAGE gel of untreated and PNGase F-treated purified Per/ROM1 complex demonstrates a shift in peripherin but not ROM1. B, staining of PVDF membranes containing purified Per/ROM1 complex with various fluorescein-conjugated lectins. C, representative MS/MS spectrum of de-glycopeptide QLTNNSAHY after PNGase F treatment. D, list of glycans detected for Asn-229 by mass spectrometry. Abbreviations: Hex, hexose; Man, mannose; HexNAc, N-acetylhexosamine; GlcNAc, N-acetylglucosamine; NeuAc, N-acetylneuraminic acid. E, sequence alignment of bovine peripherin and ROM1, transmembrane domains are underlined with broad black lines and numbered TM1–4. Peripherin glycosylation site Asn-229 is highlighted in red. F, peripherin and ROM1 two-dimensional structures displaying topology within the membrane, Asn-229 is highlighted in the peripherin structure. Red residues: missense mutation or in-frame deletion. Abbreviations: LCA, Lens culinaris agglutinin; GSLI, Griffonia (Bandeiraea) simplicifolia lectin I; PSA, Pisum sativum agglutinin; SuWGA, succinylated WGA; PHA-E, Phaseolus vulgaris agglutinin E; PHA-L, P. vulgaris agglutinin L; ConA, concanavalin A; DBA, Dolichos biflorus agglutinin; SBA, soybean agglutinin; UEAI, Ulex europaeus agglutinin I.
