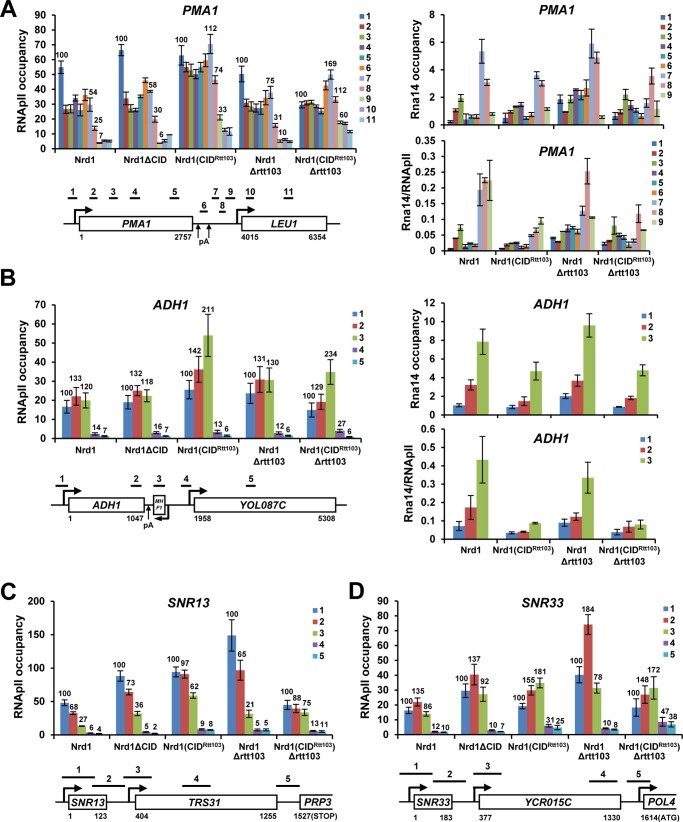
FIGURE 2.
Distinct RNApII termination profiles in Nrd1(CIDRtt103) cells. A, CID swapping of Nrd1 causes defective RNApII termination in PMA1 as measured by ChIP assay with α-Rpb3 antibody (left panel). Significant decrease of Rna14 recruitment at the 3′-end of PMA1 might result from the strong ectopic recruitment of Nrd1(CIDRtt103) to the same region and competition for CTD binding (right panel). B, CID swapping of Nrd1 does not perturb RNApII termination in ADH1 (left panel), although Rna14 recruitment was severely reduced at the 3′-end (right panel). C and D, RNApII termination begins further downstream, presumably because RNApII needs to transcribe longer where the level of Ser(P)-2 CTD becomes stronger for optimal recruitment of Nrd1(CIDRtt103). C, SNR13 gene. D, SNR33 gene. Positions of PCR primers for each geneare are illustrated. Numbers are average RNApII ChIP signals at each position (the promoter signal is set to 100). Error bars represent S.E. for at least three independent experiments.