Abstract
CYP2C22 was recently described as a retinoic acid–metabolizing cytochrome P450 enzyme whose transcription is induced by all-trans-retinoic acid (atRA) in hepatoma cells (Qian L, Zolfaghari R, and Ross AC (2010) J Lipid Res 51:1781–1792). We identified CYP2C22 as a putative nitric oxide (NO)–regulated protein in a proteomic screen and raised specific polyclonal antibodies to CYP2C22 to study its protein expression. We found that CYP2C22 is a liver-specific protein that was not significantly induced by activators of the pregnane X receptor, constitutive androstane receptor, or peroxisome proliferator-activated receptor-α, but was downregulated to <25% of control by the aryl hydrocarbon receptor agonist β-naphthoflavone in cultured rat hepatocytes. CYP2C22 protein and its mRNA both were induced by atRA in hepatocytes, with EC50 of 100–300 nM, whereas the maximal extent of mRNA induction was twice that of the protein. CYP2C22 protein, but not its mRNA, was rapidly downregulated in hepatocytes by interleukin-1 (IL-1) or NO-donating compounds, and the downregulation by IL-1 was blocked by inhibition of NO synthases. The NO donor (Z)-1-[N-(3-aminopropyl)-N-(3-ammoniopropyl)amino]diazen-1-ium-1,2-diolate reduced the half-life of CYP2C22 from 8.7 to 3.4 hours in the presence of cycloheximide, demonstrating that NO-dependent downregulation is due to stimulated proteolysis. No intermediate degradation products were detected. However, this degradation was insensitive to inhibitors of calpains or the canonical proteasomal or lysosomal pathways, indicating that NO-dependent degradation of CYP2C22 proceeds via a novel pathway.
Introduction
Nitric oxide (NO) has diverse physiologic roles, including vasodilation and blood pressure regulation (Palmer et al., 1987), neuronal communication (Dawson et al., 1992), and pathogenic cell killing (Green et al., 1990). In the latter case, NO is produced at high concentrations by the inducible NO synthase (NOS)2 (Nussler and Billiar, 1993), whose expression is induced via activation of Toll-like receptors and cytokine receptors in cells of the innate immune system as well as in hepatocytes and other cell types (Nussler and Billiar, 1993).
Activation of the immune system results in the downregulation of many drug-metabolizing enzymes (Aitken et al., 2006) and transporters (Cressman et al., 2012) in the liver, notably enzymes of the cytochrome P450 (P450) superfamily (Aitken et al., 2006). Stadler et al. (1994) first suggested that this downregulation of P450 proteins and/or mRNAs by inflammatory mediators could be attributed to NO produced via NOS2 induction. Whereas subsequent studies have revealed that this is not true for all, or even perhaps most P450s (Sewer and Morgan, 1997, 1998; Sewer et al., 1998), various studies have suggested a role for NO in the regulation of CYP2B enzymes (Khatsenko et al., 1993, 1997; Carlson and Billings, 1996). We have shown that NO produced by interleukin-1 (IL-1)β or lipopolysaccharide stimulation of hepatocytes causes ubiquitin-dependent proteasomal degradation of the rat phenobarbital-inducible P450 CYP2B1 (Ferrari et al., 2001; Lee et al., 2008; Sun et al., 2012). The related human enzyme CYP2B6 also undergoes downregulation at the protein level in response to NO (Aitken et al., 2008).
The mechanism whereby NO triggers the degradation of CYP2B1 is unknown. Both iron regulatory protein 2 (Kim et al., 2004) and alkylguanine DNA-alkyltransferase (Wei et al., 2010) undergo proteasomal degradation triggered by S-nitrosylation of critical cysteines. Although CYP2B1 can be nitrosylated by NO donors in vitro (Lee et al., 2008), changes in nitrosylation state of the protein have not been demonstrated in a cellular context. As another approach, we have been searching for other P450 enzymes that undergo NO-dependent degradation. We described earlier the isobaric tags for relative and absolute quantitation–based proteomic approach to this problem, and we used it to identify rat CYP3A enzymes as additional NO targets (Lee et al., 2009). The primary objective of the current study was to investigate the regulation by NO of an additional P450 enzyme identified in the same proteomic study, CYP2C22.
During the course of the current work, CYP2C22 was identified as a putative vitamin A–regulated gene by microarray analysis of livers from vitamin A–sufficient and –deficient rats (McClintick et al., 2006). Qian et al. (2010) replicated this observation using in situ hybridization, and they demonstrated via reverse-transcriptase polymerase chain reaction (PCR) that CYP2C22 mRNA is expressed predominantly in liver. CYP2C22 mRNA was induced by all-trans-retinoic acid (atRA) and the synthetic retinoic acid receptor (RAR) agonist Am580 in H-4-II-E rat hepatoma cells (Qian et al., 2010). Transient transfection of luciferase reporter constructs was used to identify a retinoic acid response element in the distal promoter region of the CYP2C22 gene. Moreover, CYP2C22 expression in HEK293T cells conferred higher retinoic acid–metabolizing activity upon those cells (Qian et al., 2010), identifying CYP2C22 as a retinoic acid hydroxylase. Therefore, a second objective of our work was to confirm the regulation of CYP2C22 by retinoids at the protein expression level and to investigate the regulation of this enzyme by classic inducers of hepatic P450s.
In this article, we describe the preparation of specific polyclonal antibodies to CYP2C22 and use them to investigate the regulation of this poorly understood enzyme by nitric oxide, IL-1β, and atRA, as well as by prototypical nuclear receptor agonists.
Materials and Methods
IL-1β was purchased from R&D Systems (Minneapolis, MN), and (Z)-1-[N-(3-aminopropyl)-N-(3-ammoniopropyl)amino]diazen-1-ium-1,2-diolate (DPTA NONOate, DPTA) and 3,3-bis(aminoethyl)-1-hydroxy-2-oxo-1-triazene (NOC18) were from Cayman Chemical (Ann Arbor, MI). l-NG-Nitroarginine methyl ester (l-NAME), chloroquine, Williams’ medium E, Krebs-Ringer buffer, collagenase, atRA, protease inhibitor cocktail, and other general chemicals were acquired from Sigma-Aldrich (St. Louis, MO). NADPH was from Calbiochem (Darmstadt, Germany), and brefeldin A, N-acetyl-Leu-Leu-methionine-aldehyde (ALLM), 2S,3S-trans-(ethoxycarbonyloxirane-2-carbonyl)-l-leucine-(3-methylbutyl) amide (E64d), and Z-Val-Phe-aldehyde (MDL-28170) were obtained from Enzo Life Sciences (Farmingdale, NY). Proteasome inhibitors carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132) and bortezomib were purchased from Boston Biochemicals (Cambridge, MA) and LC Laboratories (Woburn, MA), respectively. The calpain substrate 7-amino-4-chloromethylcoumarin, t-BOC-l-leucyl-l-methionine amide (Boc-LM-CMAC), was purchased from Invitrogen (Carlsbad, CA). The 4-hydroxy retinoic acid (4-OH-RA) was synthesized as described previously (Lutz et al., 2009; Shimshoni et al., 2012). Anti-CYP2B1 was kindly provided by Dr. James Halpert (University of California, San Diego, CA). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was acquired from Millipore (Billerica, MA), and anti-NOS2 and anti-annexin VI antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Anti-CYP2C11 antibody was generated in our laboratory, as described previously (Morgan et al., 1985).
Antibody Production and Peptide-Blocking Experiment.
Peptide synthesis and antibody production were carried out by Open Biosystems, Inc. (Huntsville, AL). The CYP2C22-specific peptide corresponds to residues 366–390 (24 amino acids, PRKTTQDVEFRGYHIPKGTSVMAC). Antibodies were raised in two rabbits, which were bled to measure antibody production after each injection. After the third injection of the peptide, a final bleed was performed, and then CYP2C22 antisera were prepared. To determine the specificity of the CYP2C22 antibody, various amounts of the immunogenic CYP2C22-specific peptide were immobilized by adsorption on nitrocellulose paper, which was then blocked with 5% nonfat dry milk for 1 hour. After washing three times with washing buffer (10 mM potassium phosphate, pH 7.4, 0.05% Tween 20, 1.15% KCl, 0.02% sodium azide), 5 ml of 1000-fold diluted polyclonal antibody sera was incubated overnight with the nitrocellulose paper at 4°C to immobilize CYP2C22-specific antibody. Unbound antibody solution was then taken for Western blotting to determine the specificity of antibody.
Rat Hepatocyte Isolation, Treatment, and Harvesting.
Rat hepatocytes were isolated by a two-step in situ collagenase perfusion procedure (Lee et al., 2008) with minor modifications, which are described below. The procedure was approved by the Emory University Institutional Animal Care and Use Committee. Male F344 rats (200–250 g) were anesthetized with ketamine-xylazine solution, and then the liver was perfused with 300–400 ml of Krebs-Ringer bicarbonate buffer for 8–10 minutes. This was followed by perfusion with 0.3 mg/ml collagenase type IV (Sigma-Aldrich) for 10 minutes. The liver was harvested and gently chopped to release cells, which were filtered through 90- and 200-μm meshes. Hepatocytes were cultured under conditions and treated at time points that yield optimal constitutive (Liddle et al., 1992) and phenobarbital-inducible (Sidhu et al., 1993) cytochrome P450 expression. Cells were plated on collagen plates with plating medium containing 10% fetal bovine serum, and 3 hours later, cells were overlaid with new media containing Matrigel (0.234 mg/ml; BD Biosciences, San Jose, CA). After 24 hours, the medium was changed to Williams’ medium E containing 10 mM HEPES, pH 7.4, 10 nM insulin, 25 nM dexamethasone, and penicillin/streptomycin (100 U/ml and 100 μg/ml, respectively). Cultures were maintained for 5–6 days at 37°C in 5% CO2 with regular changes in media. At the conclusion of the experiment, media were removed and reserved for a NO assay using the Griess reaction (Green et al., 1982). The cells were harvested with a cell scraper and then incubated on ice at 4°C in phosphate-buffered saline with 1 mM EDTA for at least 20 minutes to remove Matrigel. For preparation of total cell lysates, cells were pelleted at 1000g for 5 minutes at 4°C and then lysed by the addition of ice-cold 50 mM Tris-Cl buffer, pH 7.5, containing 0.1% SDS, 0.5% Nonidet P-40, 1 mM EDTA, and a protease inhibitor mixture and sonication for 10 seconds. After centrifugation for 10 minutes at 11,000g at 4°C, the supernatant was used for SDS-polyacrylamide gel electrophoresis and immunoblotting.
Preparation of Rat Liver S9 and Microsomal Fractions, and Immunoblotting.
Rat livers were homogenized in ice-cold homogenization buffer [0.1 M potassium phosphate buffer, pH 7.4, containing 0.125 M potassium chloride, 1.0 mM EDTA, and a protease inhibitor mixture [aprotinin, bestatin, trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane, l-trans-3-carboxyoxiran-2-carbonyl-l-leucylagmatine, N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide, leupeptin, and pepstatin) (Sigma P83400)] and then centrifuged at 10,000g at 4°C for 20 minutes to obtain the S9 fraction. For preparation of microsomes, the S9 fraction was centrifuged at 160,000g for 45 minutes at 4°C. The supernatant from ultracentrifugation was considered the cytosolic fraction, and the pellet was washed with cold microsome storage buffer (0.25 M sucrose containing protease inhibitor mixture) and homogenized in the same buffer.
SDS-PAGE and Western blotting were carried out as described by Lee et al. (2008). Equal amounts of each sample, typically 30–40 μg of protein, were loaded on the gels. Typically, Western analysis was carried out on freshly prepared lysates, although occasional experiments were done on lysates stored at −20°C. In our experience, lysate proteins are stable in the presence of the protease inhibitor cocktail, even after storage overnight at room temperature. Each experiment was performed using cells from a single animal, with at least three separate cell incubation replicates per treatment group. Primary antibodies (rat CYP2C22 antibody, diluted 1:3000; anti-GAPDH, 1:10,000 dilution; anti-annexin VI, 1:3000) were incubated overnight at 4°C, the blots were washed, and then horseradish peroxidase–conjugated goat anti-rabbit IgG was incubated for 1 hour at room temperature. Chemiluminescence was detected with ECL substrate (Thermo Scientific, Rockford, IL) on X-ray film. All assays were performed within a linear range, and the intensity of stained bands was measured by Bio-Rad imaging software (Image Laboratory version 4.0; Bio-Rad, Hercules, CA).
Stable Expression of CYP2C22 in HuH7 Cells.
CYP2C22 cDNA (kindly provided by Dr. A. Catherine Ross, Pennsylvania State University, University Park, PA) (Qian et al., 2010) was cloned into pLV-CMV-GFP-U3Nhe by replacing the green fluorescent protein gene, and the new construct was named Lv-CMV-2C22. Lentivirus was generated with a second generation vector system, as described by Naldini et al. (1996). Lv-CMV-2C22 or pLV-CMV-GFP-U3Nhe along with pCMVΔR8.92 and pVSVG plasmids was transiently transfected into Hek293T cells using FuGENE HD (Promega, Madison, WI). Transfection efficiency was assessed by fluorescence microscopy of green fluorescent protein expression. The lentiviruses were harvested 48 and 72 hours after transfection. The media containing virus were filtered through 0.45-μm cellulose acetate filters, and the viruses were used to transduce HuH7 cells, as described (www.addgene.org/tools/protocols/plko/). Expression of CYP2C22 protein was determined by immunoblotting.
RNA Extraction and Reverse-Transcriptase Real-Time PCR.
Total RNA was extracted with RNA-Bee reagent (Tel-Test, Friendswood, TX), according to the manufacturer's instructions. One microgram of total RNA was used for cDNA synthesis with the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA), and real-time PCR was carried out with SYBR Green PCR Master Mix (Applied Biosystems) using Eppendorf Mastercycler Realplex PCR equipment (Hauppaugge, NJ). Each reaction contained the equivalent of 50 ng of reverse-transcribed RNA. Primers were annealed at 60°C, and primer extension was for 30 seconds at 72°C for each cycle. The primers used are as follows: CYP2C11 (5′-GAATTCGCATGGATCCAGCCCTAGTCC-3′, 5′-GTCGACAATCAGATGAGAGCTTAG-3′), CYP2C22 (5′-TCACACCGGCTACCAACCCT-3′, 5′-CTGTGGGTCATGGAGAGCTG-3′), GAPDH (5′-TGCCAAGTATGATGACATCAAGAAG-3′, 5′-AGCCCAGGATGCCCTTTAGT-3′), CYP26B1 (5′-TTGAGGGCTTGGAGTTGGT-3′, 5′-AACGTTGCCATACTTCTCGC-3′), and CYP26A1 (5′-GTGCCAGTGATTGCTGAAGA-3′, 5′-GGAGGTGTCCTCTGGATGAA-3′). Melting curves were routinely checked to ensure a single PCR product. Relative expression of mRNAs was calculated by the ΔΔCt method, as described by Livak and Schmittgen (2001). GAPDH mRNA was used as the normalization control.
Hepatocyte Retinoic Acid Metabolism.
All incubations were done under red light and were conducted as duplicates of each of the biologic triplicates in the treatment group. The incubations contained 60 μg of rat hepatocyte S9 protein, 1 mM NADPH, and 2 μM atRA in 100 μl of 100 mM potassium phosphate buffer (pH 7.4). After 5 minutes at 37°C, the reactions were initiated by the addition of NADPH and allowed to proceed at 37°C for 20 minutes. A control incubation without NADPH was conducted for each group to assure that 4-OH-RA formation was NADPH dependent. The reactions were terminated with 100 μl of ice-cold acetonitrile containing the internal standard (13-cis-4oxo-RA-d3; Toronto Research Chemicals, ON, Canada).
The formation of 4-OH-RA from atRA was measured using an AB Sciex API 5500 QTrap (Foster City, CA) mass spectrometer equipped with an Agilent UHPLC 1290 (Palo Alto, CA) and Agilent Zorbax Eclipse Plus C18 (3.5 μm, 2.1 × 100 mm) column heated to 40°C. The analytes were separated using a flow rate of 300 μl/min and the following linear gradient and a mobile phase combination of water (A) and acetonitrile (B): 0 minutes, 90% A, 10% B; 3 minutes, 90% A, 10% B; 3.5 minutes, 50% A, 50% B; 7.5 minutes, 15% A, 85% B; 7.6 minutes, 5% A, 95% B; 10 minutes, 5% A, 95% B; 10.1 minutes, 90% A, 10% B; 11.5 minutes, 90% A, 10% B. The 4-OH-RA and the internal standard were detected using negative ion atmospheric pressure chemical ionization and m/z transitions of 315.1→253.2 for 4-OH-RA and 316.0→272.0 for 13cis-4oxo-RA-d3. The compound-specific mass spectrometry/mass spectrometry parameters for 4-OH-RA were DP, −40; EP, −8; CE, −23; and CXP, −19; and for 13-cis-4oxo-RA-d3 DP, −15; EP, −10; CE, −10; and CXP, −5. The data were analyzed using Analyst software, and the 4-OH-RA peak area was normalized to the peak area of the 13-cis-4oxo-RA-d3 internal standard. The percent activity remaining of 4-OH-RA formation in each incubation was calculated by comparing with the control treatment group.
Cellular Calpain Activity Assay.
Calpain activity was measured using a fluorescence-based assay with Boc-LM-CMAC as substrate (Rosser et al., 1993). Assay buffer (1 mM Na2HPO4, 137 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 2 mM CaCl2, 10 mM glucose, and 10 mM HEPES, pH 7.4) (Donato et al., 2004) containing Boc-LM-CMAC (25 μM) was added to fully confluent CYP2C22 lentivirus-transduced HuH7 cells grown in 12-well plates and incubated for 20 minutes. CMAC released from the cell due to calpain cleavage of the substrate was measured in the media with a fluorometer (excitation λ, 380 nm; emission λ, 455 nm).
Statistical Analyses.
Data are expressed as the mean ± S.E.M. Differences between groups were calculated either by t test or by one-way analysis of variance (ANOVA), followed by Dunnett’s test or Tukey’s test, as appropriate for each experimental design.
Results
Discovery of CYP2C22 as a Putative NO-Regulated Protein.
We described previously how we used isobaric tags for relative and absolute quantitation proteomic technology to identify proteins that were rapidly downregulated in a NO-dependent manner by IL-1β in cultured rat hepatocytes (Lee et al., 2009). From 229 total proteins identified, three were predicted to be regulated by NO. Two of them, CYP2B1/2 and CYP3A1/2, have been demonstrated to undergo NO-dependent proteasomal degradation in response to IL-1 and NO donors (Lee et al., 2008, 2009). The third protein was CYP2C22, which was identified from peptides KLPPGPTPLPIFGNILQVGVK and RFSLMVLR, having average abundances that were 1.9-fold lower in the samples exposed to NO (data not shown).
CYP2C22 Antibodies and Their Characterization.
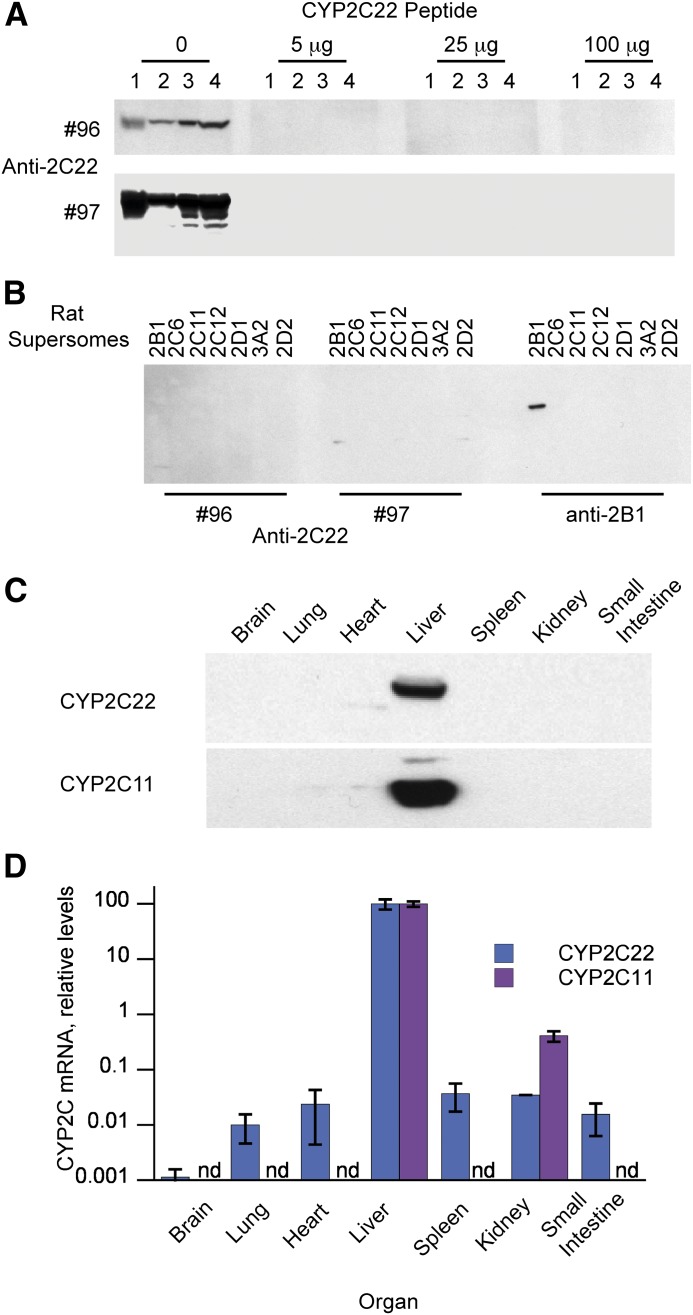
As discussed earlier, CYP2C22 has been described as a RAR-regulated mRNA in rat liver in vivo and in rat hepatoma cells, although nothing is known about its protein expression. Alignment of the amino acid sequence of CYP2C22 with those of other rat CYP2C subfamily members revealed the region from amino acids 366 to 390 to be one of greatest divergence, and therefore, this peptide was used to raise antibodies in rabbits. As shown in Fig. 1A, the antibodies from rabbit 96 recognized a single protein band in rat hepatocyte lysates, rat liver S9 fraction, and rat liver microsomes, and this was abolished by preincubation of the serum with the peptide antigen bound to nitrocellulose paper. Antibodies from rabbit 97 recognized the same band, and upon overexposure two minor bands of lower molecular weight. Preincubation of 97 serum with the antigen peptide abolished all of the bands (even longer exposure revealed one faint band of the same molecular weight as CYP2C22, which was only seen in liver S9 fractions). The 97 antibody was selected for use going forward, as it had a higher titer. The 96 and 97 antibodies failed to recognize rat CYP2B1, 2C6, 2C11, 2C12, 2D1, 2D2, or 3A2 (Fig. 1B), or human CYPs 1A1, 1A2, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, or 3A4 (data not shown). Together, these experiments establish the specificity of the antibodies for CYP2C22 in rat liver microsomes and hepatocyte lysates.
Fig. 1.
CYP2C22 antibody specificity and liver-specific expression of the enzyme. Polyclonal antibodies were raised to a peptide containing amino acids 366–190 of CYP2C22. (A) Western blots of various hepatic fractions incubated with a 1:3000 dilution of anti-CYP2C22 serum 96 or a 1:3000 dilution of antiserum 97. Before the addition to the blot, the diluted antisera were incubated overnight at 4°C with the indicated amounts of antigenic peptide adsorbed to nitrocellulose paper. Lane 1, total cell extract from primary rat hepatocytes (34 μg); lane 2, rat liver S9 fraction (40 μg); lanes 3 and 4, rat liver microsomes (5 and 15 μg, respectively). (B) Western blots using CYP2C22 antisera 96 (1:1000 dilution) or 97 (1:3000 dilution) or anti-CYP2B1 (1:5000 dilution) against insect cell microsomes expressing rat P450 enzymes (1.5-μg protein), as indicated. (C) Western blot using CYP2C22 antiserum 97 (1:3000 dilution) against microsomes from various tissues isolated from a 230-g male Sprague-Dawley rat. This blot is representative of three blots from different animals. (D) CYP2C22 and 2C11 mRNA expression in the same tissues described in (C), measured by reverse-transcriptase quantitative PCR. Relative mRNA expression is expressed as a percentage of the liver values that were arbitrarily set at 100. Values are means ± S.E.M. of values from three different animals.
Tissue-Specific Expression.
Whereas CYP2C22 mRNA was reported to be restricted to rat liver [with very low-level expression in brain (Qian et al., 2010)], the expression of CYP2C22 protein is not known. Western blotting revealed that CYP2C22 protein expression is restricted to the liver, with undetectable levels in brain, lung, heart, spleen, kidney, or small intestine (Fig. 1C); mRNA expression in these tissues was at least 1000-fold lower than liver (Fig. 1D). CYP2C11 expression, included for comparison, was also liver-specific.
Regulation by atRA.
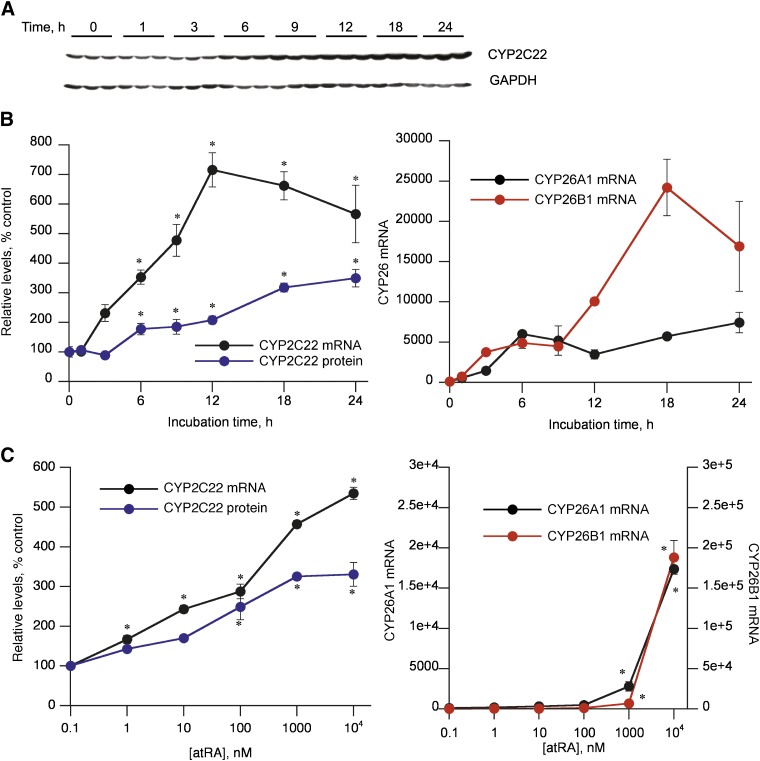
CYP2C22 protein was significantly induced 1.8-fold within 6 h of 5 μM atRA treatment of cultured hepatocytes, reaching 3.2- to 3.5-fold of control at 18–24 hours of incubation (Fig. 2, A and B). CYP2C22 mRNA was rapidly induced by atRA, by 3.6-fold at 6 hours and reaching a maximum of 7.4-fold within 12 hours (Fig. 2B). The induction of CYP2C22 protein lagged only slightly behind of its mRNA. atRA induced CYP2C22 protein with an approximate EC50 in the 10–100 nM range (Fig. 2C). CYP2C22 mRNA was induced by atRA in a similar concentration range. Induction of the mRNA for the known RAR target genes CYP26B1 and CYP26A1 had similar kinetics to those of CYP2C22, although their maximal extent of induction was much higher at 480- and 74-fold, respectively (Fig. 2B). Induction of the CYP26 mRNAs was only significant at atRA concentrations >1 μM (Fig. 2C). Again, the magnitude of CYP26B1 induction was ~10-fold higher than that of CYP26A1.
Fig. 2.
Regulation of CYP2C22 mRNA and protein by atRA in cultured rat hepatocytes. Hepatocytes were cultured for 4 days before the addition of atRA. (A) Western blot showing relative revels of CYP2C22 protein in hepatocytes treated with atRA. atRA (5 μM) was added to the cells at different times so that all samples were harvested at the same time. Control cells (0 hour) were untreated and harvested at the same time as the treated cells. (B) Quantitative analysis of the data in (A) and levels of CYP2C22 and CYP26 mRNAs in cells from the same experiment (n = 3 independent samples). *Significantly different from 0-hour control group, P < 0.05, Dunnett’s test. (C) Concentration dependencies of CYP22C and CYP2C26 induction. Hepatocytes were incubated with the indicated concentration of atRA for 24 hours. The points on the y-axis indicate cells that were treated with vehicle (dimethylsulfoxide) only. *Significantly different from untreated control group, P < 0.05, Dunnett’s test.
Regulation by Nuclear Receptor Agonists.
We next asked whether CYP2C22 expression is regulated by any of the xenobiotic receptors that regulate many of the other drug-metabolizing P450s. Cultured hepatocytes were incubated for 3 days with 40 μM β-naphthoflavone [an aryl hydrocarbon receptor (AhR) agonist], 1 mM phenobarbital [a constitutive androstane receptor activator], 20 μM dexamethasone [a pregnane X receptor (PXR) agonist], 25 μM pregnenolone 16α-carbonitrile (PCN, a PXR agonist), or 10 μM clofibrate (a peroxisome proliferator-activated receptor α agonist). We used concentrations that we and others determined previously were sufficient to induce prototypic target genes in rat hepatocytes.
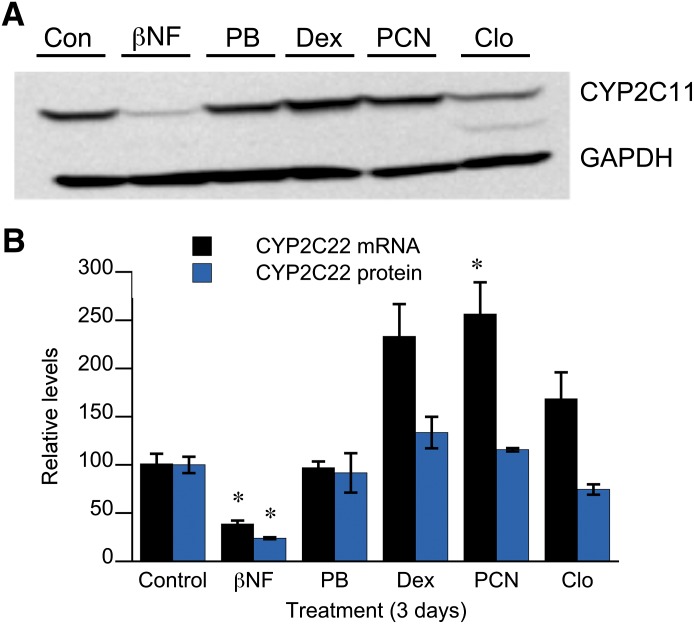
PCN caused significant induction of CYP2C22 mRNA after 3 days, whereas dexamethasone and clofibrate trended in the same direction. However, none of these agents affected the levels of CYP2C22 protein (Fig. 3). Phenobarbital had no effect on CYP2C22 mRNA or protein, whereas β-naphthoflavone downregulated both mRNA and protein to 38 and 24% of control levels, respectively (Fig. 3). The effects of β-naphthoflavone and clofibrate on CYP2C22 protein were further investigated with respect to time course (results not shown). The downregulation by β-naphthoflavone was already manifested after 24 hours of treatment, whereas clofibrate produced a small (12%) but significant suppression at 3 days only.
Fig. 3.
Effect of prototypic inducers of drug-metabolizing enzymes on CYP2C22 expression. Rat hepatocytes in sandwich cultures were treated with media (Con), 40 μM β-napthoflavone (βNF), 1 mM phenobarbital (PB), 20 μM dexamethasone (Dex), 25 μM PCN, or 10 μM clofibrate (Clo) for 72 hours, after which the cells were harvested and analyzed for CYP2C22 mRNA and protein expression. (A) Western blot of representative samples from each treatment group. (B) Quantitative analysis of individual samples (n = 3). Values are expressed relative to the control group. *Significantly different from control, P < 0.05, Dunnett’s test.
Downregulation of CYP2C22 by IL-1β via NO Generation.
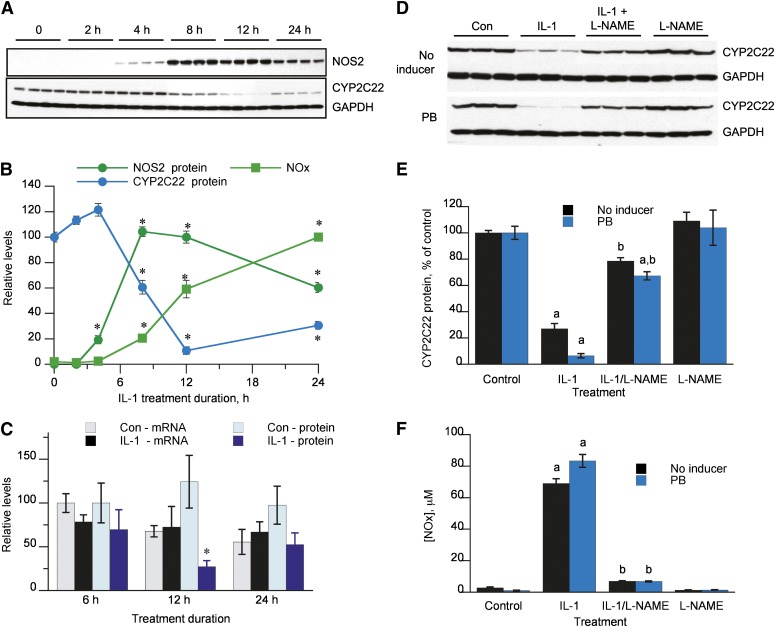
Incubation of rat hepatocytes with 5 ng/ml IL-1β resulted in a downregulation of CYP2C22 protein, which became significant within 8 hours of treatment and reached a nadir at 12 hours (Fig. 4, A and B). This corresponded with the time course of induction of NOS2 and the detection of nitric oxide products [nitrate + nitrite (NOx)] in the culture medium. Notably, NOS2 expression and the rate of NOx production declined significantly between 12 and 24 hours after treatment, during which time CYP2C22 protein expression began to recover. The rapid decline in CYP2C22 protein expression occurred in the absence of any significant effect of IL-1β on CYP2C22 mRNA levels (Fig. 4C).
Fig. 4.
NO-dependent downregulation of CYP2C22 protein. (A) Time courses of regulation of CYP2C22 and NOS2 proteins in hepatocytes treated with 5 ng/ml IL-1. (B) Quantitative analysis of the data in (A). Levels of NOx in the media are also included. *Significantly different from zero time control, P < 0.05, Dunnett’s test. (C) Post-transcriptional regulation of CYP2C22 by IL-1. Hepatocytes were treated with 5 ng/ml IL-1 and harvested at the indicated times for measurement of CYP2C22 mRNA and protein levels. *Significantly different from control cells at same time point, P < 0.05, t test. (D) Inhibition of CYP2C22 protein downregulation by l-NAME. Hepatocytes were incubated for 12 hours with control medium (Con), IL-1 (5 ng/ml), 100 μM l-NAME, or both. In the lower panel, cells were treated with 1 mM phenobarbital (PB) for 48 hours prior to and during incubation with IL-1. (E) Quantitative analysis of the data in (D) (n = 3). a, Significantly different from control; b, significantly different from IL-1–treated cells; P < 0.05, ANOVA and Tukey’s test. (F) Levels of NOx in the media of the treated cells from (D) and (E).
The NOS inhibitor l-NAME effectively blocked IL-1β–evoked NO production in the hepatocytes, and at the same time blocked the downregulation of CYP2C22 (Fig. 4, D–F). We performed this experiment with and without phenobarbital pretreatment of the cells, because our proteomic experiment in which we identified CYP2C22 as a putative NO-regulated protein was performed in phenobarbital-treated cells. The same results were seen regardless of the pretreatment (Fig. 4, D–F).
A NO Donor Mimics the Effect of IL-1β and Inhibits Retinoic Acid Metabolism in Hepatocytes.
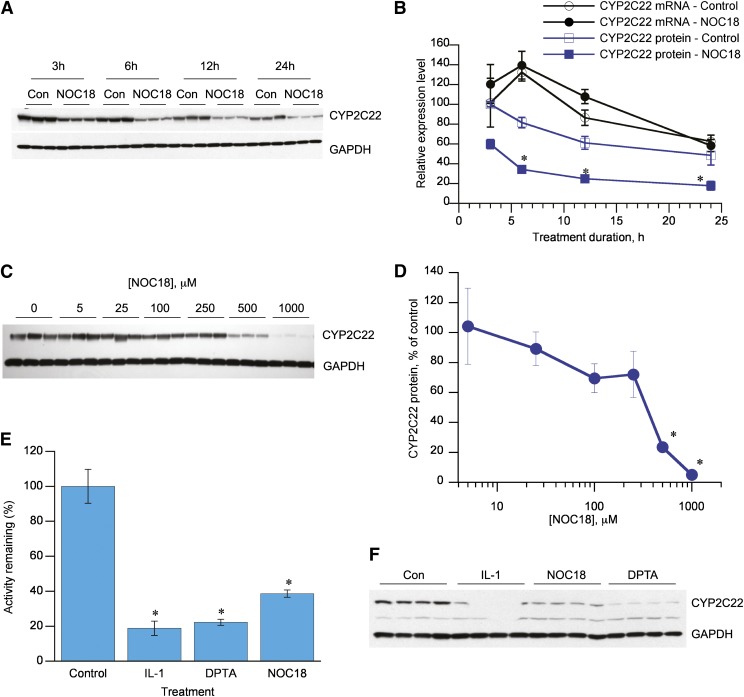
NOC18 is a NO donor that releases NO with a half-life of approximately 20 hours at 37°C (Hrabie et al., 1993), which we have demonstrated to cause the downregulation of CYP2B and CYP3A proteins in rat hepatocytes (Lee et al., 2008, 2009). The time courses of the effects of NOC18 on CYP2C22 mRNA and protein were investigated. Whereas levels of both CYP2C22 mRNA and protein changed over time in the control cells, NOC18 caused a marked downregulation of CYP2C22 protein at every time point studied (3–24 hours), while producing no effect on the corresponding mRNA levels (Fig. 5, A and B). Almost complete suppression of CYP2C22 was achieved with 1 mM NOC-18, and the IC50 was approximately 300–500 μM (Fig. 5, C and D).
Fig. 5.
Effects of NO donors on CYP2C22 expression and retinoid metabolism in hepatocytes. (A) Western blot showing time course of downregulation of CYP2C22 protein by the NO donor NOC18 in cultured hepatocytes. (B) Time courses of downregulation of CYP2C22 mRNA and protein by NOC18. Cells were treated with 500 μM NOC18 or vehicle and harvested at the indicated times. CYP2C22 data are from the blots shown in (A). *Significantly different from timed control, P < 0.05 (n = 3), t test. (C) Western blot showing concentration dependence of the NOC18 effect. Cells were treated for 12 hours with the indicated concentration. (D) Quantitative analysis of the data in (C). *Significantly different from zero time control, P < 0.05, Dunnett’s test (n = 3). (E) Effect of NO donors and IL-1 on retinoic acid–metabolizing activities of rat hepatocytes. Hepatocytes were treated with medium (control), 5 ng/ml IL-1 for 12 hours, 500 μM NOC18 for 12 hours, or 500 μM DPTA for 6 hours. The cells were harvested, and the formation of 4-OH-RA from atRA was measured in S9 fractions of the cells. The activities are shown relative to control cells. *Significantly different from control, P < 0.05, Dunnett’s test (n = 4). (F) Western blot showing CYP2C22 expression in the samples from (E).
Because CYP2C22 is a retinoic acid–metabolizing enzyme in rat liver, we next determined the effect of NO donor treatment on the retinoic acid–metabolizing activities of hepatocytes. Cells were treated with 5 ng/ml IL-1β or 500 μM NOC18 for 12 hours, or 500 μM DPTA for 6 hours, and then the cells were harvested and S9 fractions were prepared. The treatments decreased atRA 4-hydroxylation by 60 to 80%, indicating significantly decreased CYP2C22 activity (Fig. 5E). The efficacies of these treatments on CYP2C22 protein levels were verified by Western blotting (Fig. 5F), and the declines in CYP2C22 protein contents of the microsomes were correlated with comparable reductions in retinoic acid metabolism (Fig. 5, E and F). The decreased CYP2C22 protein was reflected in significantly decreased atRA hydroxylation. We note a faint lower molecular weight band detected by the CYP2C22 antibody in this experiment. Its identity is unknown, and it is only detected in some experiments.
Stimulation of CYP2C22 Protein Degradation by NO Donors.
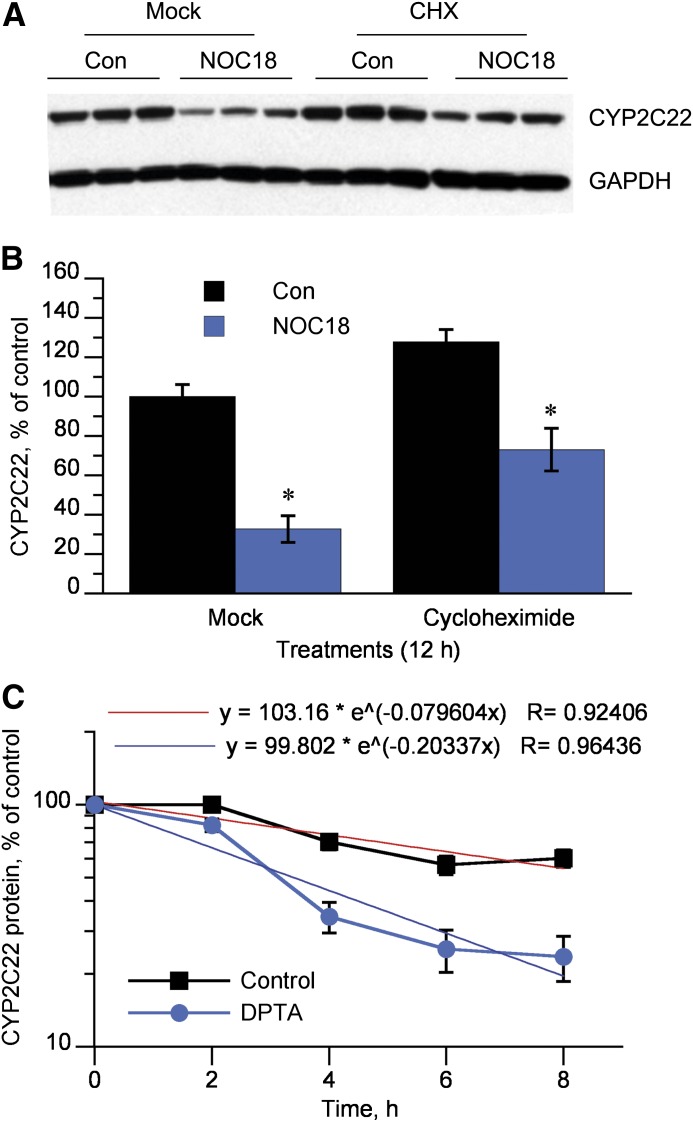
CYP2C22 protein downregulation by NOC18 was maintained in the presence of the protein synthesis inhibitor cycloheximide (Fig. 6, A and B). A time course analysis with DPTA as the NO donor showed that the DPTA reduced the half-life of CYP2C22 protein from 8.7 to 3.4 hours (Fig. 6C).
Fig. 6.
Stimulated degradation of CYP2C22 protein. (A) Western blot of lysates from cells treated for 12 hours with 500 μM NOC18 or medium (Con) in the presence (CHX) or absence (Mock) of 20 μg/ml cycloheximide. (B) Quantitative analysis of the data in (A). *Significantly different control, P < 0.05, t test (n = 3). (C) Cycloheximide chase experiment. Hepatocytes were treated with medium (Con) or 500 μM DPTA for 2, 4, 6, or 8 hours in the presence or absence of 20 μg/ml cycloheximide, and CYP2C22 levels were measured by Western blotting (n = 3). Exponential curve fitting was performed using Kaleidagraph software (Synergy Software, Reading, PA).
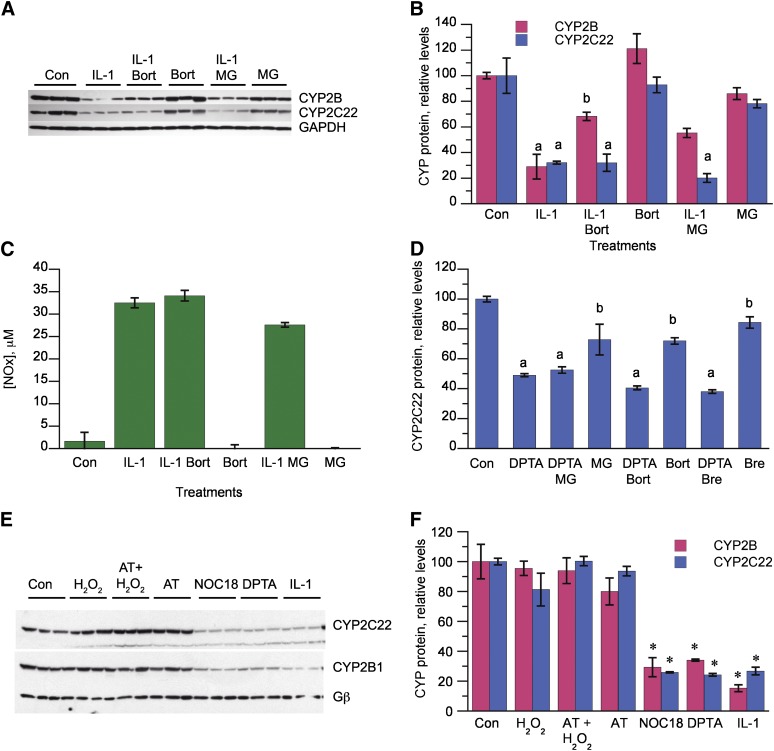
To determine the effect of proteasome inhibition on CYP2C22 downregulation, we performed the experiments in the presence of 1 mM phenobarbital to allow for expression of CYP2B1, which we have shown is degraded via the proteasome following IL-1β or NOC18 exposure of the cells. Hepatocytes were treated with the proteasome inhibitor MG132 or bortezomib 4 hours after initiation of IL-1 treatment of the cells. As we demonstrated previously, these treatments effectively blocked CYP2B1 protein downregulation (Fig. 7, A and B) while not affecting NO production by the cells (Fig. 7C). However, proteasome inhibition failed to block the downregulation of CYP2C22 (Fig. 7, A and B). We verified that proteasome inhibitors also did not block CYP2C22 downregulation by IL-1β in the absence of phenobarbital (data not shown). Proteasome inhibition also failed to block the downregulation of CYP2C22 by DPTA (Fig. 7D). Brefeldin A [inhibits endoplasmic reticulum to Golgi transport as well as endosomal trafficking (Lippincott-Schwartz et al., 1991; Strous et al., 1993)] also failed to affect CYP2C22 downregulation (Fig. 7D).
Fig. 7.
Lack of effect of proteasome, lysosome, and calpain inhibitors. (A) Western blot showing the effect of proteasome inhibitors on CYP2C22 downregulation by IL-1. Three-day-old hepatocytes were pretreated with 1 mM phenobarbital, which was present for the rest of the experiment. After 2 days of phenobarbital induction, the cells were treated with IL-1 (5 ng/ml) or control medium (Con). Four hours later, bortezomib (Bort, 10 μM), MG132 (MG, 20 μM), or control media were added. The cells were harvested 12 hours after IL-1 addition, and relative expression of CYP2C22 and CYP2B proteins was measured. (B) Quantitative analysis of the data in (A). a, Significantly different from control; b, significantly different from IL-1–treated cells; P < 0.05, ANOVA and Tukey’s test (n = 3). (C) Nitric oxide production by the cells in the experiment depicted in (A) and (B). (D) Effect of proteasome inhibitors and brefeldin A on CYP2C22 degradation stimulated by a NO donor. Hepatocytes were treated with medium (Con) or 500 μM DPTA for 5 hours in the presence or absence of bortezomib (10 μM), MG132 (20 μM), or brefeldin A (Bre, 10 μM). (E) Effect of hydrogen peroxide on CYP2C22 and CYP2B expression. Hepatocytes were pretreated with phenobarbital, as described for (A) and then treated with control medium (Con) or H2O2 (5 mM) with or without aminotriazole (AT, 25 mM) for 5 hours or with NOC18 (500 μM, 12 hours), DPTA (500 μM, 6 hours), or IL-1 (5 ng/ml, 12 hours). The cells were harvested at the same time, and the levels of CYP2C22 and CYP2B proteins were assessed by Western blotting. (F) Quantitative analysis of the data in (E). *Significantly different from control, P < 0.05, Dunnett’s test (n = 3).
In experiments not shown, we tested several different calpain and lysosomal protease inhibitors for their abilities to block the IL-1-, NOC18-, or DPTA-evoked degradation of CYP2C22 in hepatocytes at various time points and were unable to detect any significant effect. We also tested whether oxidative signals other than NO could downregulate CYP2C22 and CYP2B1. Phenobarbital-induced hepatocytes were treated with 5 mM H2O2, NO donors, or IL-1β. The NO donors and IL-1β downregulated the P450 proteins, as seen previously, but H2O2 failed to do so in the presence or absence of the catalase inhibitor aminotriazole (Fig. 7, E and F).
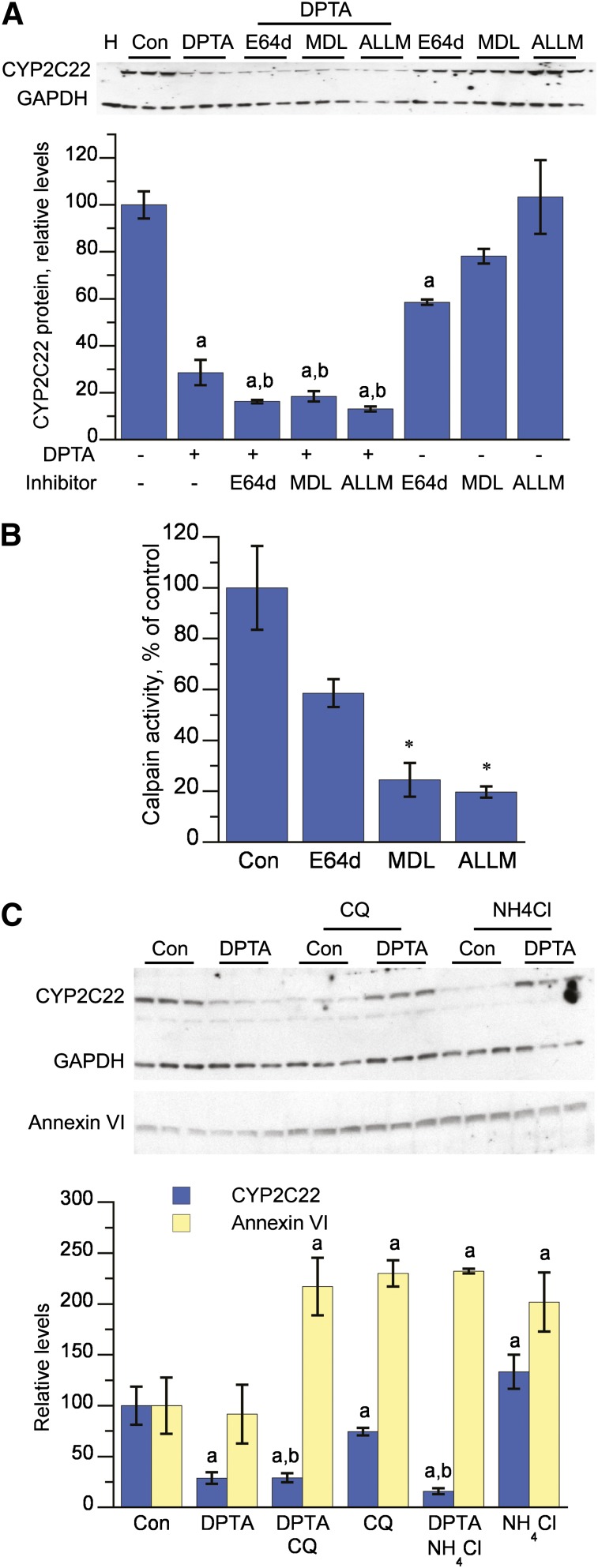
To investigate further the proteolytic enzymes responsible for CYP2C22 downregulation, we established a HuH7 cell line expressing CYP2C22 using a lentiviral vector. As shown in Fig. 8A, DPTA downregulated CYP2C22 protein expression to 30% of control in these cells, but the calpain inhibitors E64d, MDL-2180, or ALLM failed to attenuate this downregulation. MDL-2180 or ALLM effectively inhibited calpain activity, whereas E64d was less effective (Fig. 8B). The lysosome inhibitors chloroquine and NH4Cl also failed to block CYP2C22 downregulation, but effectively inhibited lysosomal degradation of annexin VI (Cuervo et al., 2000) as observed by a significant increase in annexin VI protein in the cells (Fig. 8C).
Fig. 8.
Downregulation of lentiviral-expressed CYP2C22 by NO in HuH7 cells is not attenuated by inhibitors of calpain or lysosomal degradation. (A) Effect of calpain inhibitors on CYP2C22 downregulation. Fully confluent 3-day-old HuH7 cells stably expressing CYP2C22 in 12-well plates were treated with DPTA (500 μM) and/or E64d (10 μM), MDL-28170 (MDL, 10 μM), or ALLM (10 μM) for 4 hours, and total cell lysates were harvested for CYP2C22 and GAPDH immunoblotting. a, Significantly different from control, untreated cells; b, significantly different from cells treated with inhibitor alone, P < 0.05, ANOVA and Tukey’s test (n = 3). (B) Inhibition of calpain activity. Cells cultured as described in (A) were treated with control media or calpain inhibitors E64d (10 μM), MDL-28170 (MDL, 10 μM), and ALLM (10 μM) for 2 hours. The media were exchanged for assay buffer containing the calpain substrate Boc-LM-CMAC (25 μM), and fluorescence in the media was measured after 20 minutes of incubation. *, Significantly different from control; P < 0.05, ANOVA and Dunnett’s test (n = 4). (C) Effect of lysosomotropic agents. To study lysosome-dependent degradation of annexin VI, cells were first deprived of serum for 4 hours to enhance the expression of annexin VI, and then serum was added to stimulate its degradation. DPTA (500 μM) and/or chloroquine (CQ, 100 μM) or NH4Cl (20 mM) were added concomitantly with the serum, and the cells were incubated for an additional 4 hours. Total cell lysates were harvested for Western blotting. a, Significantly different from control, untreated cells; b, significantly different from cells treated with inhibitor alone, P < 0.05, ANOVA and Tukey’s test (n = 3).
Discussion
Our data establish CYP2C22 as a third P450 protein targeted by NO-mediated degradation in rat hepatocytes, based on the prevention of IL-1β–mediated suppression by NOS inhibitors; suppression of protein expression in the absence of significant effects on the cognate mRNA; and its rapid downregulation by NO donors in the presence of the protein synthesis inhibitor cycloheximide. By use of DPTA, a NO donor with a chemical half-life in aqueous solution of 3 hours (Hrabie et al., 1993), the half-life of CYP2C22 was reduced by 60% to 3.4 hours. Whereas the kinetics of downregulation could be fitted to an exponential curve (R = 0.964), the data gave some indication of a biphasic decay (Fig. 6C). Therefore, if anything, the estimate of 3.4 hours may underestimate the initial rapid decay of the protein. Furthermore, we demonstrate that this effect is specific to NO and is not reproduced by oxidant stress caused by application of hydrogen peroxide in the presence or absence of catalase inhibition.
The downregulation of CYP2C22 by IL-1β and the NO donors DPTA and NOC18 was accompanied by similar changes in the retinoic acid hydroxylase activities of the cells, supporting the reported finding of CYP2C22 as a retinoic acid hydroxylase (Qian et al., 2010). This significant role of CYP2C22 in atRA metabolism is most likely due to the fact that CYP26 enzyme expression is low in the absence of atRA treatment as shown in human hepatocytes and HepG2 cells (Tay et al., 2010). The remaining atRA hydroxylation activity in the IL-1 and NO donor-treated hepatocytes could be attributed to the contribution of CYP26 enzymes to atRA hydroxylation.
Unlike CYP2B1 (Lee et al., 2008; Sun et al., 2012) and CYP3A (Lee et al., 2009), however, the downregulation of CYP2C22 by NO could not be inhibited by proteasome inhibitors. Lysosomal degradation is the main route of degradation of CYP2B and some other P450 enzymes in their native states (Ronis et al., 1991; Roberts, 1997; Liao et al., 2005), but we found that the lysosomal inhibitors NH4Cl and chloroquine, as well as three different calpain inhibitors, all failed to inhibit CYP2C22 downregulation. Therefore, we conclude that CYP2C22 degradation in response to NO proceeds via a novel pathway. The protein is not being trafficked to a different cellular compartment, because whole cellular lysates were used. Also, we have not detected the appearance of any lower molecular weight bands during CYP2C22 degradation, even on overexposed blots from cells treated with proteasome inhibitors.
The possibility that CYP2C22 was being cleaved in the antigenic domain (amino acids 366–390) was also investigated. In experiments not shown, we determined that a different polyclonal antibody raised to the entire CYP2C11 protein recognizes a protein in rat hepatocytes that undergoes rapid NO-dependent downregulation similar to that of CYP2C22. It has been documented that CYP2C22 expression increases when hepatocytes are cultured, whereas CYP2C11 declines (Emi et al., 1990). Because we have determined that CYP2C11 is not a NO-responsive protein (Sewer and Morgan, 1997; Lee et al., 2009), we conclude that this CYP2C11 antibody detects primarily CYP2C22 in these cultures. By use of this polyclonal CYP2C antibody, we were also unable to detect discreet cleavage products in overexposed blots from cells treated with proteasome inhibitors and NO donors. Clearly, the CYP2C11 antibody recognizes additional CYP2C epitopes to the specific CYP2C22 antibody. Therefore, we conclude it to be unlikely that the disappearance of the protein is due to a peptidase making a discreet cut between amino acids 366 and 390.
CYP2C11 and CYP2C22 share 61% amino acid identity (298 of 489), yet CYP2C22 undergoes NO-stimulated degradation, whereas CYP2C11 does not. The contrasting behaviors of these highly related proteins may provide a strategy from which to identify structural features that confer response to NO. As mentioned above, cysteine nitrosylation sensitizes iron regulatory protein 2 and alkylguanine DNA-alkyltransferase (Kim et al., 2004; Wei et al., 2010) to proteasomal degradation. In contrast, tyrosine nitration of CYP2B1 on Tyr190 is causal for its catalytic inactivation by peroxynitrite (Lin et al., 2005), and nitration could possibly contribute to CYP2C22 degradation. CYP2C22 has four cysteine residues (Cys13, Cys55, Cys164, and Cys389) and six tyrosine residues (Tyr78, Tyr151, Tyr219, Tyr223, Tyr265, and Tyr315) that are not present in CYP2C11. Human CYP2C8, 2C9, and 2C19 share 60.7, 62.0, and 61.6% amino acid identity with CYP2C22. Cys164 and Cys389 are present in all three human CYP2Cs, and Cys13 is present in CYP2C9 and 2C19. None of the six tyrosines absent from 2C11 are present in the human CYP2C enzymes. The contributions of these residues to NO-mediated degradation are currently under investigation. However, it must also be considered that larger structural features could be involved in the susceptibility to degradation.
By use of antibodies specific to CYP2C22, we demonstrated that its tissue distribution is the same as reported previously for its mRNA specificity (Qian et al., 2010). In a rat hepatoma cell line, CYP2C22 mRNA was induced approximately 10-fold by 100 nM to 1 μM atRA, whereas higher concentrations were not tested (Qian et al., 2010). Our data in primary hepatocytes are in good agreement with this finding. It is noteworthy that CYP2C22 mRNA and CYP26B1 levels began to decline between 12 and 24 hours of treatment, possibly reflecting increased atRA metabolism attributed to the induction of these proteins (A. Topletz and N. Isoherranen, unpublished data). As a consequence, CYP2C22 protein levels never reached the same magnitude of induction as CYP2C22 mRNA. Nevertheless, CYP2C22 protein began to be induced only 3 hours after the mRNA was first seen to increase, consistent with our determination of a half-life of only 8.7 hours for the protein in the absence of NO.
Westin et al. (1993, 1997) found that the closely related CYP2C7 mRNA was induced by atRA in rat hepatocytes and was partially dependent on atRA metabolism. However, significant induction was only observed at atRA concentrations >1 μM in hepatocytes (Westin et al., 1993), whereas CYP2C22 protein and mRNA induction can be detected with 100 nM atRA. Together with the data of Qian et al. (2010), this suggests that CYP2C22 induction, in contrast to CYP2C7, proceeds via a classic RAR mechanism. It is noteworthy that Qian et al. (2010) also found that human CYP2C9 is induced by atRA in human hepatocytes. This perhaps raises the question of whether CYP2C22 and CYP2C9 might share other regulatory mechanisms, such as stimulated degradation by NO.
From our experiments in cultured rat hepatocytes, CYP2C22 protein is not significantly regulated by activators of the receptors PXR, constitutive androstane receptor, or peroxisome proliferator-activated receptor α, which are important in the regulation of many xenobiotic-metabolizing P450 enzymes. Although Emi et al. (1990) concluded from qualitative analysis of Northern blots that phenobarbital and the AhR agonist 3-methylcholanthrene did not induce hepatic CYP2C22 mRNA in vivo, closer inspection of their Northern blots suggests that levels were slightly reduced by 3-methylcholanthrene. In support of this interpretation, our data show a strong and reproducible downregulation by β-naphthoflavone of CYP2C22 mRNA and protein levels in rat hepatocytes. The potent AhR agonist tetrachlorodibenzo-p-dioxin causes depletion of hepatic retinoid stores, possibly via increased metabolism by AhR-responsive P450 and UDP-glucuronosyltransferase enzymes (Novak et al., 2008). Further research will be needed to determine whether such a mechanism could explain the rapid downregulation of CYP2C22 mRNA by β-naphthoflavone.
We showed previously that, similar to rat CYP2B1, human CYP2B6 protein undergoes NO-dependent downregulation in human hepatocytes (Aitken et al., 2008). In addition to CYP2C22, Qian et al. (2010) showed the related human CYP2C9 mRNA to be regulated by atRA. Whether any of the human CYP2C enzymes are regulated by nitric oxide is a subject of ongoing investigation. However, until the mechanisms of NO-mediated downregulation are better understood, it is not possible to predict the susceptibility of individual forms from amino acid sequence or indeed from tertiary structures.
Acknowledgments
The authors thank Ayush Kishore (Department of Pharmacology) for preliminary data and Dr. Dennis Koop (Oregon Health Sciences University) and Dr. Leslie Dickmann (Amgen, Inc.) for helpful discussions.
Abbreviations
- AhR
aryl hydrocarbon receptor
- ALLM
N-acetyl-Leu-Leu-methionine-aldehyde
- ANOVA
analysis of variance
- atRA
all-trans-retinoic acid
- Boc-LM-CMAC
BOC-l-leucyl-l-methionine amide
- DPTA
dipropylenetriamine NONOate
- E64d
2S,3S-trans-(ethoxycarbonyloxirane-2-carbonyl)- l-leucine-(3-methylbutyl) amide
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IL-1
interleukin-1
- l-NAME
l-NG-nitroarginine methyl ester
- MDL-28170
Z-Val-Phe-aldehyde
- MG132
carbobenzoxy-l-leucyl-l-leucyl-l-leucinal
- NO
nitric oxide
- NOC18
3,3-bis(aminoethyl)-1-hydroxy-2-oxo-1-triazene
- NOS
inducible NO synthase
- NOx
nitrate + nitrite
- 4-OH-RA
4-hydroxy retinoic acid
- P450
cytochrome P450
- PCN
pregnenolone 16α-carbonitrile
- PCR
polymerase chain reaction
- PXR
pregnane X receptor
- RAR
retinoic acid receptor
Authorship Contributions
Conducted experiments: C.-m. Lee, B.-s. Lee, Arnold.
Performed data analysis: C.-m. Lee, B.-s. Lee, Arnold, Isoherranen, Morgan.
Wrote or contributed to the writing of the manuscript: C.-m. Lee, B.-s. Lee, Arnold, Isoherranen, Morgan.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Science [Grants R01 GM069971 and R01 GM081569].
References
- Aitken AE, Lee CM, Morgan ET. (2008) Roles of nitric oxide in inflammatory downregulation of human cytochromes P450. Free Radic Biol Med 44:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken AE, Richardson TA, Morgan ET. (2006) Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol 46:123–149 [DOI] [PubMed] [Google Scholar]
- Carlson TJ, Billings RE. (1996) Role of nitric oxide in the cytokine-mediated regulation of cytochrome P-450. Mol Pharmacol 49:796–801 [PubMed] [Google Scholar]
- Cressman AM, Petrovic V, Piquette-Miller M. (2012) Inflammation-mediated changes in drug transporter expression/activity: implications for therapeutic drug response. Expert Rev Clin Pharmacol 5:69–89 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Gomes AV, Barnes JA, Dice JF. (2000) Selective degradation of annexins by chaperone-mediated autophagy. J Biol Chem 275:33329–33335 [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL, Snyder SH. (1992) A novel neuronal messenger molecule in brain: the free radical, nitric oxide. Ann Neurol 32:297–311 [DOI] [PubMed] [Google Scholar]
- Donato MT, Jiménez N, Castell JV, Gómez-Lechón MJ. (2004) Fluorescence-based assays for screening nine cytochrome P450 (P450) activities in intact cells expressing individual human P450 enzymes. Drug Metab Dispos 32:699–706 [DOI] [PubMed] [Google Scholar]
- Emi Y, Chijiiwa C, Omura T. (1990) A different cytochrome P450 form is induced in primary cultures of rat hepatocytes. Proc Natl Acad Sci USA 87:9746–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari L, Peng N, Halpert JR, Morgan ET. (2001) Role of nitric oxide in down-regulation of CYP2B1 protein, but not RNA, in primary cultures of rat hepatocytes. Mol Pharmacol 60:209–216 [DOI] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. (1982) Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem 126:131–138 [DOI] [PubMed] [Google Scholar]
- Green SJ, Meltzer MS, Hibbs JB, Jr, Nacy CA. (1990) Activated macrophages destroy intracellular Leishmania major amastigotes by an L-arginine-dependent killing mechanism. J Immunol 144:278–283 [PubMed] [Google Scholar]
- Hrabie JA, Klose JR, Wink DA, Keefer LK. (1993) New nitric oxide-releasing zwitterions derived from polyamines. J Org Chem 58:1472–1476 [Google Scholar]
- Khatsenko OG, Boobis AR, Gross SS. (1997) Evidence for nitric oxide participation in down-regulation of CYP2B1/2 gene expression at the pretranslational level. Toxicol Lett 90:207–216 [DOI] [PubMed] [Google Scholar]
- Khatsenko OG, Gross SS, Rifkind AB, Vane JR. (1993) Nitric oxide is a mediator of the decrease in cytochrome P450-dependent metabolism caused by immunostimulants. Proc Natl Acad Sci USA 90:11147–11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wing SS, Ponka P. (2004) S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol Cell Biol 24:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Kim BY, Li L, Morgan ET. (2008) Nitric oxide-dependent proteasomal degradation of cytochrome P450 2B proteins. J Biol Chem 283:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Pohl J, Morgan ET. (2009) Dual mechanisms of CYP3A protein regulation by proinflammatory cytokine stimulation in primary hepatocyte cultures. Drug Metab Dispos 37:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Zgoda VG, Murray BP, Correia MA. (2005) Vacuolar degradation of rat liver CYP2B1 in Saccharomyces cerevisiae: further validation of the yeast model and structural implications for the degradation of mammalian endoplasmic reticulum P450 proteins. Mol Pharmacol 67:1460–1469 [DOI] [PubMed] [Google Scholar]
- Liddle C, Mode A, Legraverend C, Gustafsson JA. (1992) Constitutive expression and hormonal regulation of male sexually differentiated cytochromes P450 in primary cultured rat hepatocytes. Arch Biochem Biophys 298:159–166 [DOI] [PubMed] [Google Scholar]
- Lin HL, Zhang H, Waskell L, Hollenberg PF. (2005) The highly conserved Glu149 and Tyr190 residues contribute to peroxynitrite-mediated nitrotyrosine formation and the catalytic activity of cytochrome P450 2B1. Chem Res Toxicol 18:1203–1210 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. (1991) Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67:601–616 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. (2009) Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem Pharmacol 77:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Crabb DW, Tian H, Pinaire J, Smith JR, Jerome RE, Edenberg HJ. (2006) Global effects of vitamin A deficiency on gene expression in rat liver: evidence for hypoandrogenism. J Nutr Biochem 17:345–355 [DOI] [PubMed] [Google Scholar]
- Morgan ET, MacGeoch C, Gustafsson JA. (1985) Sexual differentiation of cytochrome P-450 in rat liver: evidence for a constitutive isozyme as the male-specific 16 alpha-hydroxylase. Mol Pharmacol 27:471–479 [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- Novák J, Benísek M, Hilscherová K. (2008) Disruption of retinoid transport, metabolism and signaling by environmental pollutants. Environ Int 34:898–913 [DOI] [PubMed] [Google Scholar]
- Nussler AK, Billiar TR. (1993) Inflammation, immunoregulation, and inducible nitric oxide synthase. J Leukoc Biol 54:171–178 [PubMed] [Google Scholar]
- Palmer RM, Ferrige AG, Moncada S. (1987) Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 327:524–526 [DOI] [PubMed] [Google Scholar]
- Qian L, Zolfaghari R, Ross AC. (2010) Liver-specific cytochrome P450 CYP2C22 is a direct target of retinoic acid and a retinoic acid-metabolizing enzyme in rat liver. J Lipid Res 51:1781–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BJ. (1997) Evidence of proteasome-mediated cytochrome P-450 degradation. J Biol Chem 272:9771–9778 [DOI] [PubMed] [Google Scholar]
- Ronis MJ, Johansson I, Hultenby K, Lagercrantz J, Glaumann H, Ingelman-Sundberg M. (1991) Acetone-regulated synthesis and degradation of cytochrome P450E1 and cytochrome P4502B1 in rat liver [corrected]. Eur J Biochem 198:383–389 [DOI] [PubMed] [Google Scholar]
- Rosser BG, Powers SP, Gores GJ. (1993) Calpain activity increases in hepatocytes following addition of ATP: demonstration by a novel fluorescent approach. J Biol Chem 268:23593–23600 [PubMed] [Google Scholar]
- Sewer MB, Barclay TB, Morgan ET. (1998) Down-regulation of cytochrome P450 mRNAs and proteins in mice lacking a functional NOS2 gene. Mol Pharmacol 54:273–279 [DOI] [PubMed] [Google Scholar]
- Sewer MB, Morgan ET. (1997) Nitric oxide-independent suppression of P450 2C11 expression by interleukin-1beta and endotoxin in primary rat hepatocytes. Biochem Pharmacol 54:729–737 [DOI] [PubMed] [Google Scholar]
- Sewer MB, Morgan ET. (1998) Down-regulation of the expression of three major rat liver cytochrome P450S by endotoxin in vivo occurs independently of nitric oxide production. J Pharmacol Exp Ther 287:352–358 [PubMed] [Google Scholar]
- Shimshoni JA, Roberts AG, Scian M, Topletz AR, Blankert SA, Halpert JR, Nelson WL, Isoherranen N. (2012) Stereoselective formation and metabolism of 4-hydroxy-retinoic acid enantiomers by cytochrome p450 enzymes. J Biol Chem 287:42223–42232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu JS, Farin FM, Omiecinski CJ. (1993) Influence of extracellular matrix overlay on phenobarbital-mediated induction of CYP2B1, 2B2, and 3A1 genes in primary adult rat hepatocyte culture. Arch Biochem Biophys 301:103–113 [DOI] [PubMed] [Google Scholar]
- Stadler J, Trockfeld J, Schmalix WA, Brill T, Siewert JR, Greim H, Doehmer J. (1994) Inhibition of cytochromes P4501A by nitric oxide. Proc Natl Acad Sci USA 91:3559–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous GJ, van Kerkhof P, van Meer G, Rijnboutt S, Stoorvogel W. (1993) Differential effects of brefeldin A on transport of secretory and lysosomal proteins. J Biol Chem 268:2341–2347 [PubMed] [Google Scholar]
- Sun H, Lee CM, Tripathi S, Kim KB, Morgan ET. (2012) Nitric oxide-dependent CYP2B degradation is potentiated by a cytokine-regulated pathway and utilizes the immunoproteasome subunit LMP2. Biochem J 445:377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S, Dickmann L, Dixit V, Isoherranen N. (2010) A comparison of the roles of peroxisome proliferator-activated receptor and retinoic acid receptor on CYP26 regulation. Mol Pharmacol 77:218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. (2010) S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med 2:19ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin S, Mode A, Murray M, Chen R, Gustafsson JA. (1993) Growth hormone and vitamin A induce P4502C7 mRNA expression in primary rat hepatocytes. Mol Pharmacol 44:997–1002 [PubMed] [Google Scholar]
- Westin S, Sonneveld E, van der Leede BM, van der Saag PT, Gustafsson JA, Mode A. (1997) CYP2C7 expression in rat liver and hepatocytes: regulation by retinoids. Mol Cell Endocrinol 129:169–179 [DOI] [PubMed] [Google Scholar]