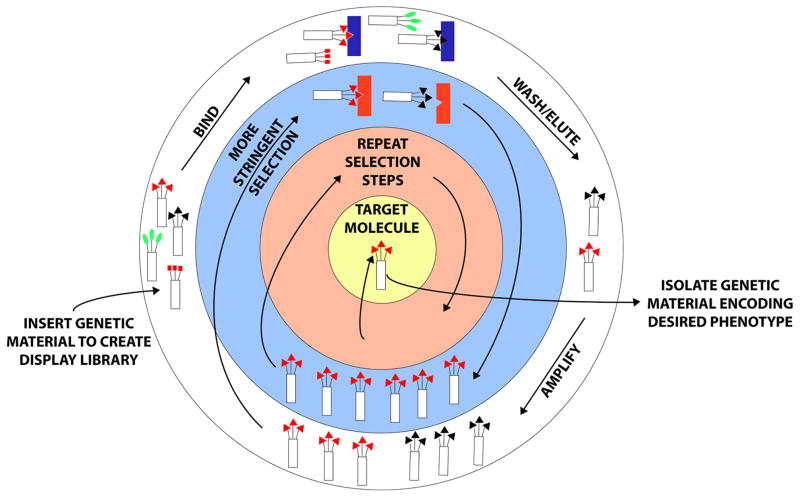
Figure 1.
Molecular Display. A protein or peptide with specific properties can be derived via molecular display following multiple rounds of selection. Polypeptide libraries can be made or purchased and either inserted into molecular hosts such as phage or yeast or appended to carrier molecules such as mRNA or ribosomes. The schematic above is representative of phage or cell-surface display, where proteins are displayed on the surfaces of organisms that are subsequently exposed to various substrates, which serve as selection criteria. In the example above, protein-mediated binding of the organism with a specific substrate followed by washing away of non-specific binders and elution enables amplification of those organisms displaying proteins with the desired properties. Repetitive screening with additional or more stringent selection criteria enables ultimate isolation of an optimized protein sequence based on phenotype, which can then be linked with genotype following extraction of genetic material from the organism. Thus, molecular display facilitates discovery of information needed to design a protein with specific properties evolved to match nearly any chosen selection criteria.