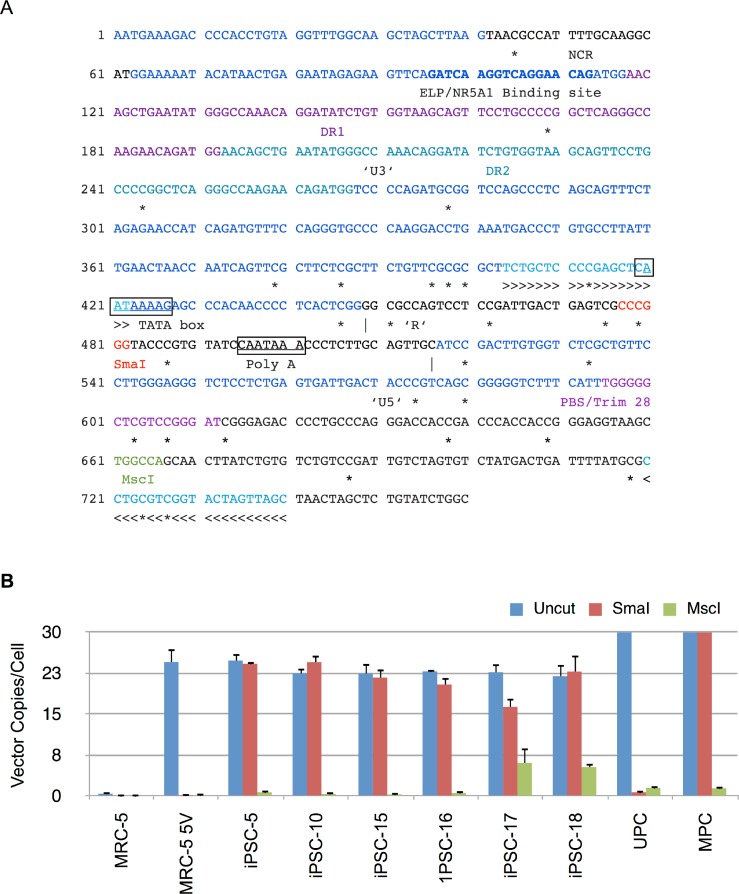
Figure 8. Methylation analysis of Moloney vector 5′ LTR using MSRE-qPCR.
(A) Sequence of 5′ long terminal repeat (LTR) and untranslated region of Moloney murine leukemia virus. The U3, R and U5 sequences within the LTR are shown and demarcated by vertical lines. Also shown are direct repeats (DR1 and DR2), Tata box, polyadenylation signal (Poly A), negative control region (NCR), binding site for ELP/NR5A1, and primer binding site (PBS). The CpG nucleotides are marked underneath by ‘*’ to indicate putative sites of methylation. The methylation sensitive SmaI and methylation insensitive MscI restriction enzyme sites are shown in red and green, respectively. >>> and <<< identify forward and reverse primers used in MSRE-qPCR. (B) Genomic DNA from untransduced MRC-5, vector transduced MRC-5 cells (MRC-5 5V), iPSC clones, and MRC-5 control genomic DNA spiked with unmethylated (UPC) or methylated plasmid vector DNA (MPC) was either undigested (Uncut) or digested with SmaI or MscI and then subjected to qPCR for determination of vector 5′ LTR or β-Actin copy numbers. The vector copy numbers per cell was calculated as described in Materials and Methods. Error bars represent one standard deviation. The standard deviation of the ratio of means was calculated as described under Materials and Methods.